hd_model_test() validates the model on new data. It takes an already tuned model,
evaluates it on the validation (new test) set, calculates the metrics and plots the probability
and ROC curve based on the new data.
Usage
hd_model_test(
model_object,
train_set,
test_set,
variable = "Disease",
metadata_cols = NULL,
case,
control = NULL,
balance_groups = TRUE,
palette = NULL,
seed = 123
)Arguments
- model_object
An
hd_modelobject coming fromhd_model_rreg()andhd_model_rf()binary or multiclass classification.- train_set
The training set as an HDAnalyzeR object or a dataset in wide format with sample ID as its first column and class column as its second column.
- test_set
The validation/test set as an HDAnalyzeR object or a dataset in wide format with sample ID as its first column and class column as its second column.
- variable
The name of the metadata variable containing the case and control groups. Default is "Disease".
- metadata_cols
The metadata variables to include in the analysis. Default is NULL.
- case
The case class.
- control
The control groups. If NULL, it will be set to all other unique values of the variable that are not the case. Default is NULL.
- balance_groups
Whether to balance the groups in the train set. It is only valid in binary classification settings. Default is TRUE.
- palette
The color palette for the classes. If it is a character, it should be one of the palettes from
hd_palettes(). Default is NULL.- seed
Seed for reproducibility. Default is 123.
Value
The model object containing the validation set, the metrics, the ROC curve, the probability plot, and the confusion matrix for the new data.
Details
In order to run this function, the train and test sets should be in exactly
the same format meaning that they must have the same columns in the same order.
Some function arguments like the case/control, variable, and metadata_cols should
be also the same. If the data contain missing values, KNN (k=5) imputation
will be used to impute. If case is provided, the model will be a binary
classification model. If case is NULL, the model will be a multiclass classification model.
In multi-class models, the groups in the train set are not balanced and sensitivity and specificity are calculated via macro-averaging. In case the model is run against a continuous variable, the palette will be ignored.
Examples
# Initialize an HDAnalyzeR object
hd_object <- hd_initialize(example_data, example_metadata)
# Split the data for training and validation sets
dat <- hd_object$data
train_indices <- sample(1:nrow(dat), size = floor(0.8 * nrow(dat)))
train_data <- dat[train_indices, ]
validation_data <- dat[-train_indices, ]
hd_object_train <- hd_initialize(train_data, example_metadata, is_wide = TRUE)
hd_object_val <- hd_initialize(validation_data, example_metadata, is_wide = TRUE)
# Split the training set into training and inner test sets
hd_split <- hd_split_data(hd_object_train, variable = "Disease")
#> Warning: Too little data to stratify.
#> • Resampling will be unstratified.
# Run the regularized regression model pipeline
model_object <- hd_model_rreg(hd_split,
variable = "Disease",
case = "AML",
grid_size = 5,
palette = "cancers12",
verbose = FALSE)
#> The groups in the train set are balanced. If you do not want to balance the groups, set `balance_groups = FALSE`.
# Run the model evaluation pipeline
hd_model_test(model_object, hd_object_train, hd_object_val, case = "AML", palette = "cancers12")
#> The groups in the train set are balanced. If you do not want to balance the groups, set `balance_groups = FALSE`.
#> $train_data
#> # A tibble: 66 × 102
#> DAid Disease AARSD1 ABL1 ACAA1 ACAN ACE2 ACOX1 ACP5 ACP6
#> <chr> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 DA00006 1 6.83 1.18e+0 -1.74 -0.156 1.53 -0.721 0.620 0.527
#> 2 DA00049 1 4.48 4.56e+0 4.86 0.230 2.24 2.97 2.60 -1.11
#> 3 DA00023 1 2.92 -7.06e-5 0.602 1.59 0.198 1.61 0.283 2.35
#> 4 DA00041 1 2.19 1.66e+0 -0.0167 -0.567 3.77 0.369 1.38 1.09
#> 5 DA00029 1 4.04 1.41e+0 -2.09 0.427 0.200 0.537 0.0262 0.105
#> 6 DA00020 1 1.80 1.70e+0 2.77 -1.04 1.33 -0.0247 1.02 0.112
#> 7 DA00045 1 2.99 2.24e+0 -0.180 -0.00102 0.367 0.604 0.843 -1.96
#> 8 DA00046 1 3.03 3.90e-1 1.83 0.983 2.60 0.113 0.504 1.42
#> 9 DA00002 1 1.42 1.25e+0 -0.816 -0.459 0.826 -0.902 0.647 1.30
#> 10 DA00011 1 3.48 4.96e+0 3.50 -0.338 4.48 1.26 2.18 1.62
#> # ℹ 56 more rows
#> # ℹ 92 more variables: ACTA2 <dbl>, ACTN4 <dbl>, ACY1 <dbl>, ADA <dbl>,
#> # ADA2 <dbl>, ADAM15 <dbl>, ADAM23 <dbl>, ADAM8 <dbl>, ADAMTS13 <dbl>,
#> # ADAMTS15 <dbl>, ADAMTS16 <dbl>, ADAMTS8 <dbl>, ADCYAP1R1 <dbl>,
#> # ADGRE2 <dbl>, ADGRE5 <dbl>, ADGRG1 <dbl>, ADGRG2 <dbl>, ADH4 <dbl>,
#> # ADM <dbl>, AGER <dbl>, AGR2 <dbl>, AGR3 <dbl>, AGRN <dbl>, AGRP <dbl>,
#> # AGXT <dbl>, AHCY <dbl>, AHSP <dbl>, AIF1 <dbl>, AIFM1 <dbl>, AK1 <dbl>, …
#>
#> $test_data
#> # A tibble: 117 × 102
#> DAid Disease AARSD1 ABL1 ACAA1 ACAN ACE2 ACOX1 ACP5 ACP6
#> <chr> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 DA00402 0 2.47 0.793 -0.996 0.790 0.0125 -0.946 0.122 2.25
#> 2 DA00331 0 2.54 1.74 5.07 0.378 NA 0.295 2.26 1.71
#> 3 DA00072 0 3.78 2.58 2.01 0.241 0.168 1.47 1.04 0.925
#> 4 DA00315 0 3.66 1.71 0.812 0.690 -0.618 1.50 1.42 2.62
#> 5 DA00476 0 3.64 2.66 NA -0.0880 NA 2.53 1.94 0.634
#> 6 DA00239 0 2.97 2.73 2.66 1.03 0.471 0.636 -0.600 1.88
#> 7 DA00340 0 2.85 0.719 0.918 2.64 3.18 0.342 1.72 -2.39
#> 8 DA00059 0 4.29 1.66 4.13 1.56 3.66 -1.06 2.09 1.26
#> 9 DA00413 0 3.08 2.53 0.426 NA 0.339 NA 2.17 2.10
#> 10 DA00203 0 3.51 0.603 1.86 0.00641 0.143 0.587 -0.383 2.00
#> # ℹ 107 more rows
#> # ℹ 92 more variables: ACTA2 <dbl>, ACTN4 <dbl>, ACY1 <dbl>, ADA <dbl>,
#> # ADA2 <dbl>, ADAM15 <dbl>, ADAM23 <dbl>, ADAM8 <dbl>, ADAMTS13 <dbl>,
#> # ADAMTS15 <dbl>, ADAMTS16 <dbl>, ADAMTS8 <dbl>, ADCYAP1R1 <dbl>,
#> # ADGRE2 <dbl>, ADGRE5 <dbl>, ADGRG1 <dbl>, ADGRG2 <dbl>, ADH4 <dbl>,
#> # ADM <dbl>, AGER <dbl>, AGR2 <dbl>, AGR3 <dbl>, AGRN <dbl>, AGRP <dbl>,
#> # AGXT <dbl>, AHCY <dbl>, AHSP <dbl>, AIF1 <dbl>, AIFM1 <dbl>, AK1 <dbl>, …
#>
#> $model_type
#> [1] "binary_class"
#>
#> $final_workflow
#> ══ Workflow ════════════════════════════════════════════════════════════════════
#> Preprocessor: Recipe
#> Model: logistic_reg()
#>
#> ── Preprocessor ────────────────────────────────────────────────────────────────
#> 5 Recipe Steps
#>
#> • step_dummy()
#> • step_nzv()
#> • step_normalize()
#> • step_corr()
#> • step_impute_knn()
#>
#> ── Model ───────────────────────────────────────────────────────────────────────
#> Logistic Regression Model Specification (classification)
#>
#> Main Arguments:
#> penalty = 4.32976659024308e-06
#> mixture = 0.307220224633347
#>
#> Computational engine: glmnet
#>
#>
#> $metrics
#> $metrics$accuracy
#> [1] 0.7606838
#>
#> $metrics$sensitivity
#> [1] 0.875
#>
#> $metrics$specificity
#> [1] 0.7522936
#>
#> $metrics$auc
#> [1] 0.940367
#>
#> $metrics$confusion_matrix
#> Truth
#> Prediction 0 1
#> 0 82 1
#> 1 27 7
#>
#>
#> $roc_curve
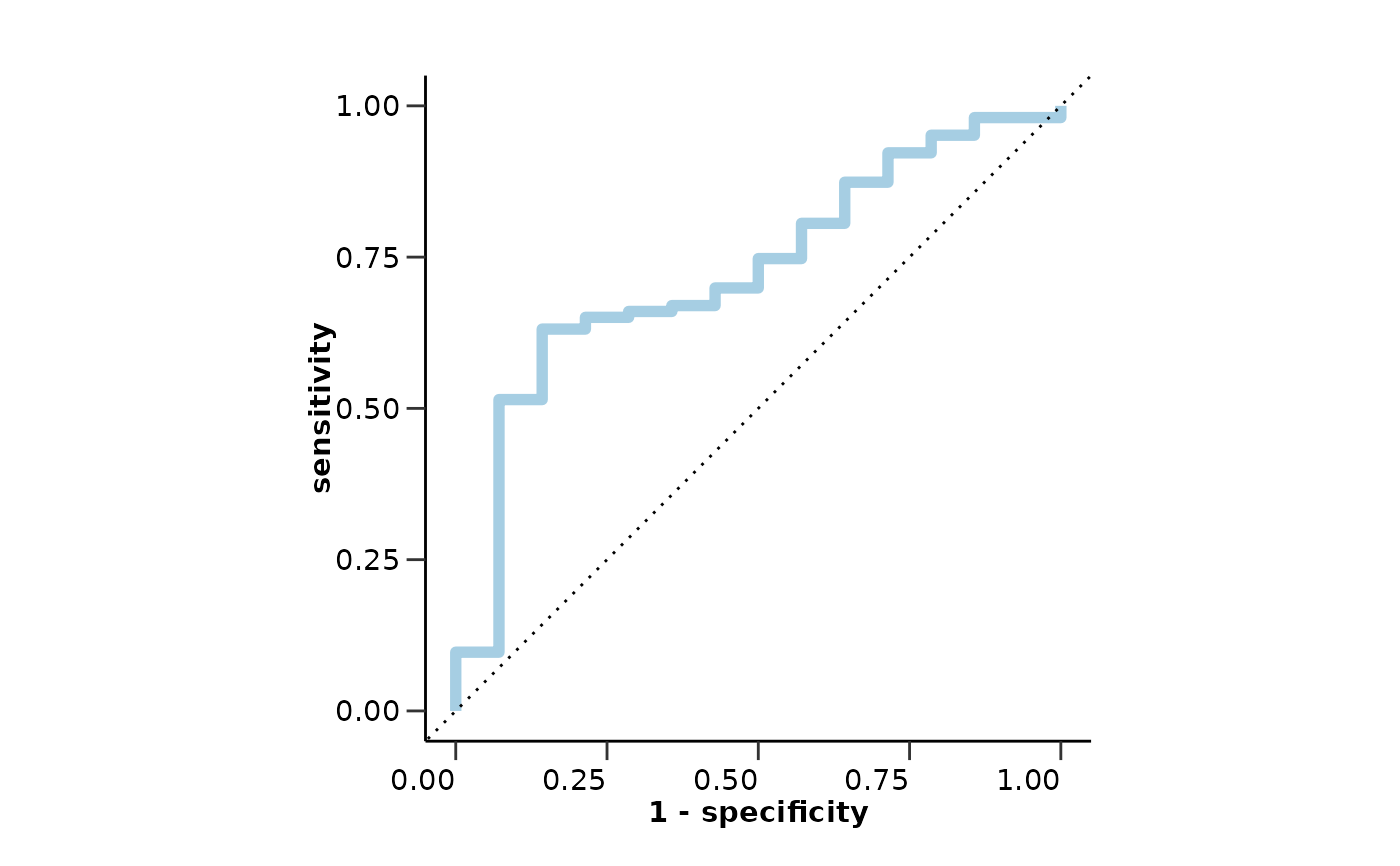 #>
#> $probability_plot
#>
#> $probability_plot
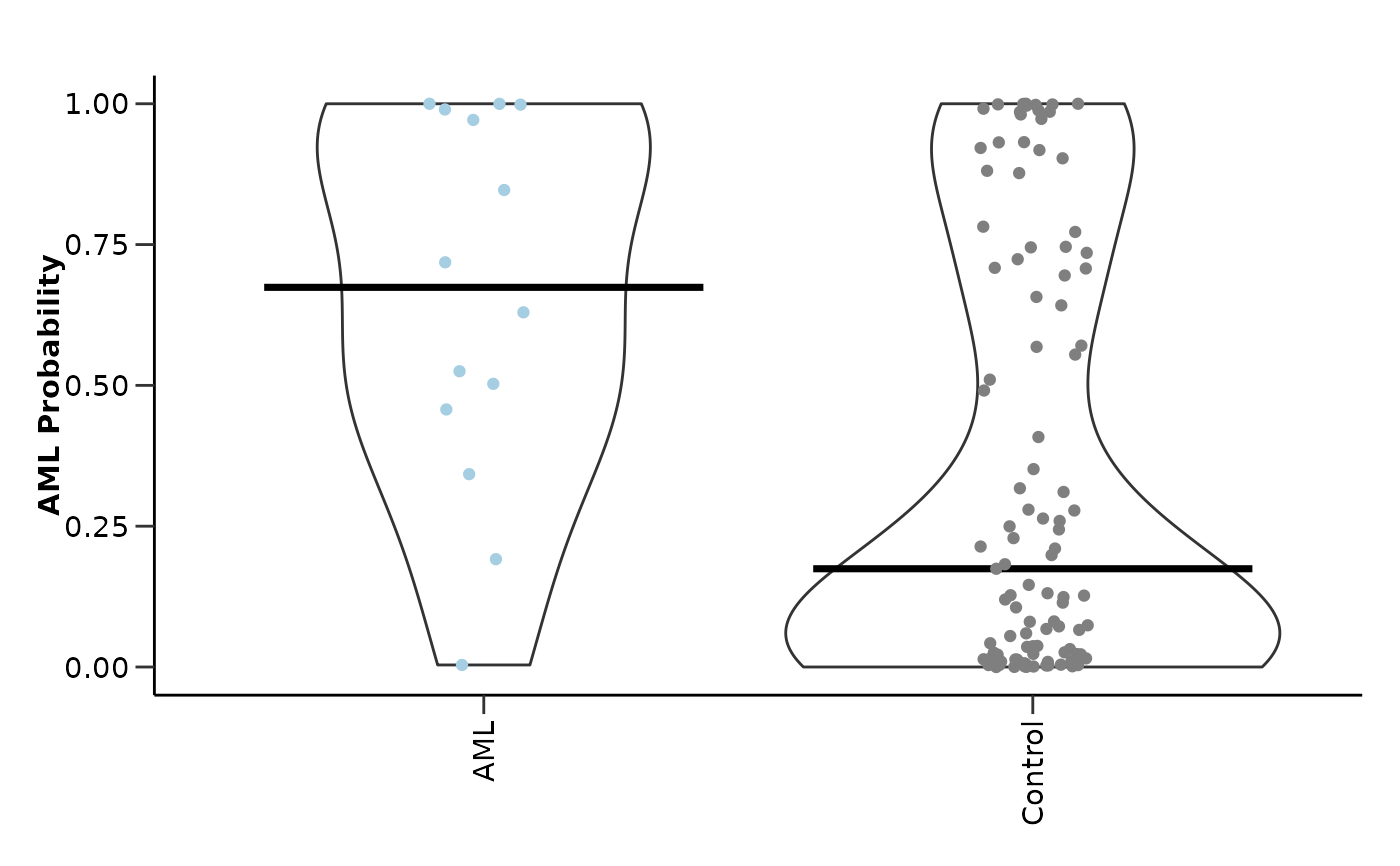 #>
#> $mixture
#> [1] 0.3072202
#>
#> $features
#> # A tibble: 100 × 4
#> Feature Importance Sign Scaled_Importance
#> <fct> <dbl> <chr> <dbl>
#> 1 ANGPT1 1.06 NEG 1
#> 2 AK1 0.988 NEG 0.929
#> 3 ALPP 0.844 NEG 0.793
#> 4 AHCY 0.819 POS 0.770
#> 5 ADAM8 0.646 NEG 0.607
#> 6 AIFM1 0.604 POS 0.567
#> 7 AXIN1 0.584 NEG 0.549
#> 8 AKT1S1 0.529 POS 0.497
#> 9 ARID4B 0.522 NEG 0.491
#> 10 APEX1 0.521 POS 0.490
#> # ℹ 90 more rows
#>
#> $feat_imp_plot
#>
#> $mixture
#> [1] 0.3072202
#>
#> $features
#> # A tibble: 100 × 4
#> Feature Importance Sign Scaled_Importance
#> <fct> <dbl> <chr> <dbl>
#> 1 ANGPT1 1.06 NEG 1
#> 2 AK1 0.988 NEG 0.929
#> 3 ALPP 0.844 NEG 0.793
#> 4 AHCY 0.819 POS 0.770
#> 5 ADAM8 0.646 NEG 0.607
#> 6 AIFM1 0.604 POS 0.567
#> 7 AXIN1 0.584 NEG 0.549
#> 8 AKT1S1 0.529 POS 0.497
#> 9 ARID4B 0.522 NEG 0.491
#> 10 APEX1 0.521 POS 0.490
#> # ℹ 90 more rows
#>
#> $feat_imp_plot
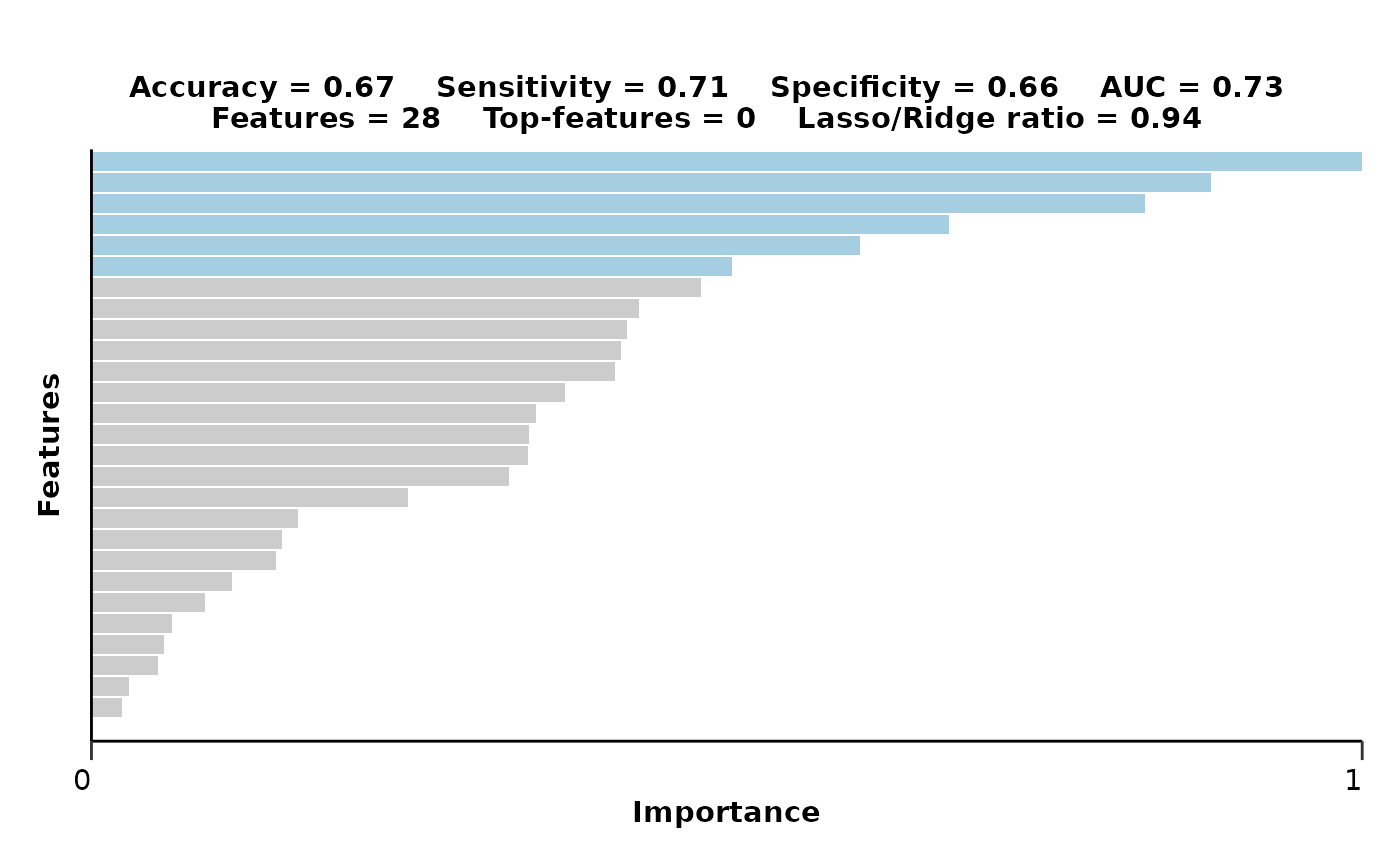 #>
#> $validation_data
#> # A tibble: 118 × 102
#> DAid Disease AARSD1 ABL1 ACAA1 ACAN ACE2 ACOX1 ACP5 ACP6
#> <chr> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 DA00003 1 NA NA NA 0.989 NA 0.330 1.37 NA
#> 2 DA00007 1 NA NA 3.96 0.682 3.14 2.62 1.47 2.25
#> 3 DA00012 1 4.31 0.710 -1.44 -0.218 -0.469 -0.361 -0.0714 -1.30
#> 4 DA00014 1 6.34 7.25 5.12 0.0193 1.29 0.370 -0.382 0.830
#> 5 DA00016 1 1.79 1.36 0.106 -0.372 3.40 -1.19 1.77 1.07
#> 6 DA00030 1 3.31 5.38 4.82 0.266 0.606 3.12 1.22 2.13
#> 7 DA00032 1 3.62 3.06 -1.34 0.965 1.05 1.53 0.152 -0.124
#> 8 DA00038 1 2.23 1.42 0.484 1.72 1.46 0.0747 1.82 0.109
#> 9 DA00043 1 2.48 1.49 0.605 0.339 0.436 0.690 1.11 0.0158
#> 10 DA00051 0 2.53 3.00 0.166 0.707 -0.00699 1.05 0.898 1.53
#> # ℹ 108 more rows
#> # ℹ 92 more variables: ACTA2 <dbl>, ACTN4 <dbl>, ACY1 <dbl>, ADA <dbl>,
#> # ADA2 <dbl>, ADAM15 <dbl>, ADAM23 <dbl>, ADAM8 <dbl>, ADAMTS13 <dbl>,
#> # ADAMTS15 <dbl>, ADAMTS16 <dbl>, ADAMTS8 <dbl>, ADCYAP1R1 <dbl>,
#> # ADGRE2 <dbl>, ADGRE5 <dbl>, ADGRG1 <dbl>, ADGRG2 <dbl>, ADH4 <dbl>,
#> # ADM <dbl>, AGER <dbl>, AGR2 <dbl>, AGR3 <dbl>, AGRN <dbl>, AGRP <dbl>,
#> # AGXT <dbl>, AHCY <dbl>, AHSP <dbl>, AIF1 <dbl>, AIFM1 <dbl>, AK1 <dbl>, …
#>
#> $test_metrics
#> $test_metrics$accuracy
#> [1] 0.6779661
#>
#> $test_metrics$sensitivity
#> [1] 0.8888889
#>
#> $test_metrics$specificity
#> [1] 0.6605505
#>
#> $test_metrics$auc
#> [1] 0.8980632
#>
#> $test_metrics$confusion_matrix
#> Truth
#> Prediction 0 1
#> 0 72 1
#> 1 37 8
#>
#>
#> $test_roc_curve
#>
#> $validation_data
#> # A tibble: 118 × 102
#> DAid Disease AARSD1 ABL1 ACAA1 ACAN ACE2 ACOX1 ACP5 ACP6
#> <chr> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 DA00003 1 NA NA NA 0.989 NA 0.330 1.37 NA
#> 2 DA00007 1 NA NA 3.96 0.682 3.14 2.62 1.47 2.25
#> 3 DA00012 1 4.31 0.710 -1.44 -0.218 -0.469 -0.361 -0.0714 -1.30
#> 4 DA00014 1 6.34 7.25 5.12 0.0193 1.29 0.370 -0.382 0.830
#> 5 DA00016 1 1.79 1.36 0.106 -0.372 3.40 -1.19 1.77 1.07
#> 6 DA00030 1 3.31 5.38 4.82 0.266 0.606 3.12 1.22 2.13
#> 7 DA00032 1 3.62 3.06 -1.34 0.965 1.05 1.53 0.152 -0.124
#> 8 DA00038 1 2.23 1.42 0.484 1.72 1.46 0.0747 1.82 0.109
#> 9 DA00043 1 2.48 1.49 0.605 0.339 0.436 0.690 1.11 0.0158
#> 10 DA00051 0 2.53 3.00 0.166 0.707 -0.00699 1.05 0.898 1.53
#> # ℹ 108 more rows
#> # ℹ 92 more variables: ACTA2 <dbl>, ACTN4 <dbl>, ACY1 <dbl>, ADA <dbl>,
#> # ADA2 <dbl>, ADAM15 <dbl>, ADAM23 <dbl>, ADAM8 <dbl>, ADAMTS13 <dbl>,
#> # ADAMTS15 <dbl>, ADAMTS16 <dbl>, ADAMTS8 <dbl>, ADCYAP1R1 <dbl>,
#> # ADGRE2 <dbl>, ADGRE5 <dbl>, ADGRG1 <dbl>, ADGRG2 <dbl>, ADH4 <dbl>,
#> # ADM <dbl>, AGER <dbl>, AGR2 <dbl>, AGR3 <dbl>, AGRN <dbl>, AGRP <dbl>,
#> # AGXT <dbl>, AHCY <dbl>, AHSP <dbl>, AIF1 <dbl>, AIFM1 <dbl>, AK1 <dbl>, …
#>
#> $test_metrics
#> $test_metrics$accuracy
#> [1] 0.6779661
#>
#> $test_metrics$sensitivity
#> [1] 0.8888889
#>
#> $test_metrics$specificity
#> [1] 0.6605505
#>
#> $test_metrics$auc
#> [1] 0.8980632
#>
#> $test_metrics$confusion_matrix
#> Truth
#> Prediction 0 1
#> 0 72 1
#> 1 37 8
#>
#>
#> $test_roc_curve
 #>
#> $test_probability_plot
#>
#> $test_probability_plot
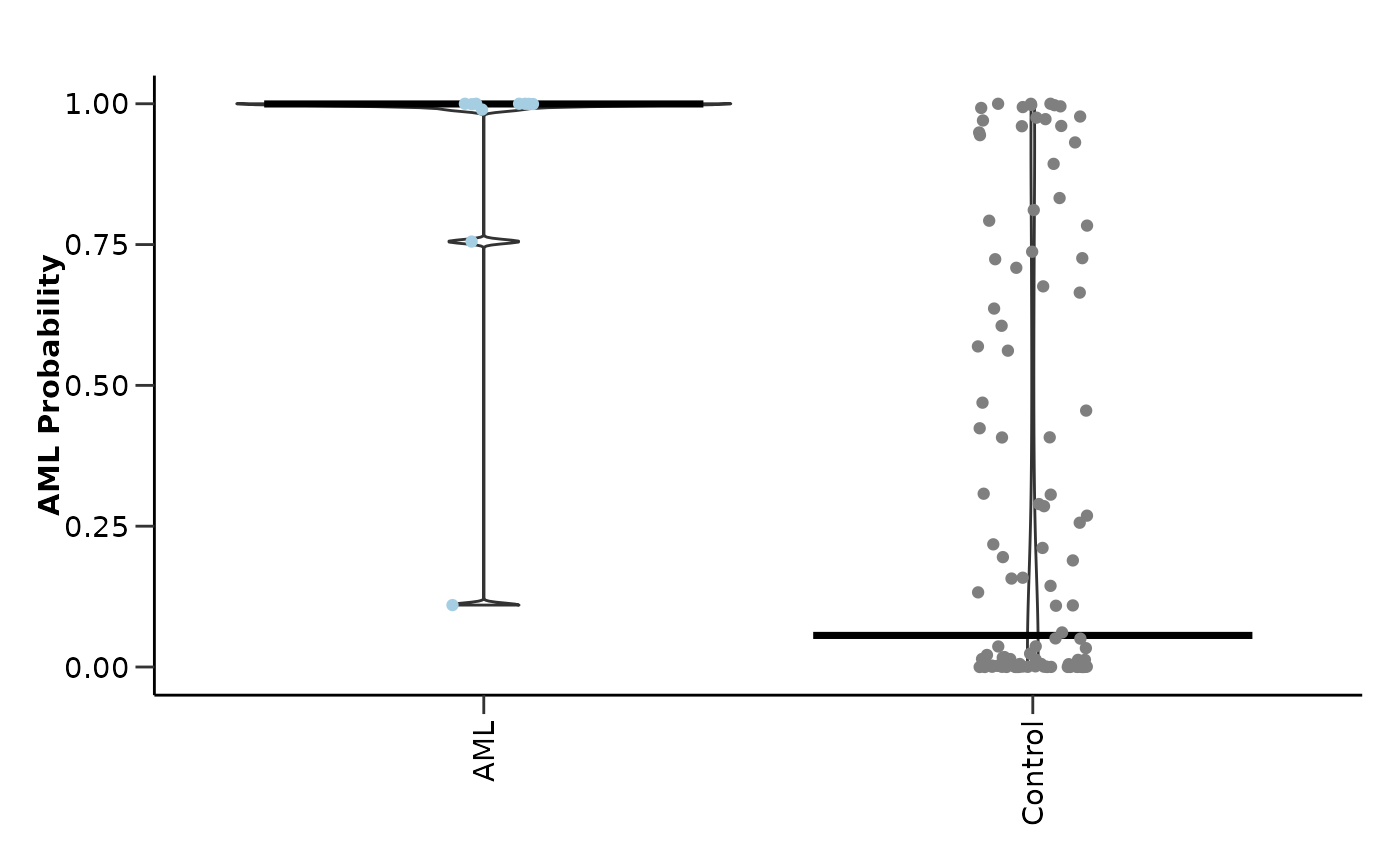 #>
#> attr(,"class")
#> [1] "hd_model"
# Run the pipeline against continuous variable
# Split the training set into training and inner test sets
hd_split <- hd_split_data(hd_object_train, variable = "Age")
# Run the regularized regression model pipeline
model_object <- hd_model_rreg(hd_split,
variable = "Age",
case = "AML",
grid_size = 2,
cv_sets = 2,
plot_title = NULL,
verbose = FALSE)
#> The groups in the train set are balanced. If you do not want to balance the groups, set `balance_groups = FALSE`.
# Run the model evaluation pipeline
hd_model_test(model_object, hd_object_train, hd_object_val, variable = "Age", case = NULL)
#> The groups in the train set are balanced. If you do not want to balance the groups, set `balance_groups = FALSE`.
#> $train_data
#> # A tibble: 350 × 102
#> DAid Age AARSD1 ABL1 ACAA1 ACAN ACE2 ACOX1 ACP5 ACP6
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 DA00315 48 3.66 1.71 0.812 0.690 -0.618 1.50 1.42 2.62
#> 2 DA00424 45 3.75 1.64 NA 0.414 1.84 1.83 0.829 2.02
#> 3 DA00049 40 4.48 4.56 4.86 0.230 2.24 2.97 2.60 -1.11
#> 4 DA00292 48 4.25 5.63 4.43 -0.467 -0.310 2.93 0.907 3.18
#> 5 DA00203 44 3.51 0.603 1.86 0.00641 0.143 0.587 -0.383 2.00
#> 6 DA00445 44 2.98 0.680 -0.310 -0.279 1.56 -0.240 -0.368 0.0101
#> 7 DA00221 51 NA NA -0.881 0.447 0.458 -0.562 0.442 0.546
#> 8 DA00229 52 2.86 3.89 3.42 1.26 0.883 2.70 1.13 2.60
#> 9 DA00023 42 2.92 -0.0000706 0.602 1.59 0.198 1.61 0.283 2.35
#> 10 DA00079 49 4.49 3.66 NA 1.85 NA 2.03 1.76 2.52
#> # ℹ 340 more rows
#> # ℹ 92 more variables: ACTA2 <dbl>, ACTN4 <dbl>, ACY1 <dbl>, ADA <dbl>,
#> # ADA2 <dbl>, ADAM15 <dbl>, ADAM23 <dbl>, ADAM8 <dbl>, ADAMTS13 <dbl>,
#> # ADAMTS15 <dbl>, ADAMTS16 <dbl>, ADAMTS8 <dbl>, ADCYAP1R1 <dbl>,
#> # ADGRE2 <dbl>, ADGRE5 <dbl>, ADGRG1 <dbl>, ADGRG2 <dbl>, ADH4 <dbl>,
#> # ADM <dbl>, AGER <dbl>, AGR2 <dbl>, AGR3 <dbl>, AGRN <dbl>, AGRP <dbl>,
#> # AGXT <dbl>, AHCY <dbl>, AHSP <dbl>, AIF1 <dbl>, AIFM1 <dbl>, AK1 <dbl>, …
#>
#> $test_data
#> # A tibble: 118 × 102
#> DAid Age AARSD1 ABL1 ACAA1 ACAN ACE2 ACOX1 ACP5 ACP6 ACTA2
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 DA004… 81 2.47 0.793 -0.996 0.790 0.0125 -0.946 0.122 2.25 3.52
#> 2 DA005… 86 NA NA 0.441 0.185 -0.234 -1.26 1.30 0.792 0.491
#> 3 DA004… 51 3.63 2.89 3.73 0.481 0.319 0.712 1.48 1.90 3.60
#> 4 DA001… 47 3.14 1.77 NA -0.489 -0.780 1.41 1.40 1.79 -0.298
#> 5 DA001… 60 6.75 7.43 1.63 0.478 0.305 0.274 1.18 3.01 1.92
#> 6 DA000… 86 6.83 1.18 -1.74 -0.156 1.53 -0.721 0.620 0.527 0.772
#> 7 DA002… 71 2.97 2.73 2.66 1.03 0.471 0.636 -0.600 1.88 1.45
#> 8 DA003… 88 2.85 0.719 0.918 2.64 3.18 0.342 1.72 -2.39 0.614
#> 9 DA001… 86 3.13 1.91 -0.296 -0.0146 0.0954 -1.02 0.378 0.735 1.23
#> 10 DA005… 81 4.32 3.08 2.44 0.617 1.12 1.29 1.31 1.72 2.20
#> # ℹ 108 more rows
#> # ℹ 91 more variables: ACTN4 <dbl>, ACY1 <dbl>, ADA <dbl>, ADA2 <dbl>,
#> # ADAM15 <dbl>, ADAM23 <dbl>, ADAM8 <dbl>, ADAMTS13 <dbl>, ADAMTS15 <dbl>,
#> # ADAMTS16 <dbl>, ADAMTS8 <dbl>, ADCYAP1R1 <dbl>, ADGRE2 <dbl>, ADGRE5 <dbl>,
#> # ADGRG1 <dbl>, ADGRG2 <dbl>, ADH4 <dbl>, ADM <dbl>, AGER <dbl>, AGR2 <dbl>,
#> # AGR3 <dbl>, AGRN <dbl>, AGRP <dbl>, AGXT <dbl>, AHCY <dbl>, AHSP <dbl>,
#> # AIF1 <dbl>, AIFM1 <dbl>, AK1 <dbl>, AKR1B1 <dbl>, AKR1C4 <dbl>, …
#>
#> $model_type
#> [1] "regression"
#>
#> $final_workflow
#> ══ Workflow ════════════════════════════════════════════════════════════════════
#> Preprocessor: Recipe
#> Model: linear_reg()
#>
#> ── Preprocessor ────────────────────────────────────────────────────────────────
#> 5 Recipe Steps
#>
#> • step_dummy()
#> • step_nzv()
#> • step_normalize()
#> • step_corr()
#> • step_impute_knn()
#>
#> ── Model ───────────────────────────────────────────────────────────────────────
#> Linear Regression Model Specification (regression)
#>
#> Main Arguments:
#> penalty = 2.60336915648323e-09
#> mixture = 0.249163427995518
#>
#> Computational engine: glmnet
#>
#>
#> $metrics
#> $metrics$rmse
#> [1] 18.17391
#>
#> $metrics$rsq
#> [1] 0.02013586
#>
#>
#> $comparison_plot
#>
#> attr(,"class")
#> [1] "hd_model"
# Run the pipeline against continuous variable
# Split the training set into training and inner test sets
hd_split <- hd_split_data(hd_object_train, variable = "Age")
# Run the regularized regression model pipeline
model_object <- hd_model_rreg(hd_split,
variable = "Age",
case = "AML",
grid_size = 2,
cv_sets = 2,
plot_title = NULL,
verbose = FALSE)
#> The groups in the train set are balanced. If you do not want to balance the groups, set `balance_groups = FALSE`.
# Run the model evaluation pipeline
hd_model_test(model_object, hd_object_train, hd_object_val, variable = "Age", case = NULL)
#> The groups in the train set are balanced. If you do not want to balance the groups, set `balance_groups = FALSE`.
#> $train_data
#> # A tibble: 350 × 102
#> DAid Age AARSD1 ABL1 ACAA1 ACAN ACE2 ACOX1 ACP5 ACP6
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 DA00315 48 3.66 1.71 0.812 0.690 -0.618 1.50 1.42 2.62
#> 2 DA00424 45 3.75 1.64 NA 0.414 1.84 1.83 0.829 2.02
#> 3 DA00049 40 4.48 4.56 4.86 0.230 2.24 2.97 2.60 -1.11
#> 4 DA00292 48 4.25 5.63 4.43 -0.467 -0.310 2.93 0.907 3.18
#> 5 DA00203 44 3.51 0.603 1.86 0.00641 0.143 0.587 -0.383 2.00
#> 6 DA00445 44 2.98 0.680 -0.310 -0.279 1.56 -0.240 -0.368 0.0101
#> 7 DA00221 51 NA NA -0.881 0.447 0.458 -0.562 0.442 0.546
#> 8 DA00229 52 2.86 3.89 3.42 1.26 0.883 2.70 1.13 2.60
#> 9 DA00023 42 2.92 -0.0000706 0.602 1.59 0.198 1.61 0.283 2.35
#> 10 DA00079 49 4.49 3.66 NA 1.85 NA 2.03 1.76 2.52
#> # ℹ 340 more rows
#> # ℹ 92 more variables: ACTA2 <dbl>, ACTN4 <dbl>, ACY1 <dbl>, ADA <dbl>,
#> # ADA2 <dbl>, ADAM15 <dbl>, ADAM23 <dbl>, ADAM8 <dbl>, ADAMTS13 <dbl>,
#> # ADAMTS15 <dbl>, ADAMTS16 <dbl>, ADAMTS8 <dbl>, ADCYAP1R1 <dbl>,
#> # ADGRE2 <dbl>, ADGRE5 <dbl>, ADGRG1 <dbl>, ADGRG2 <dbl>, ADH4 <dbl>,
#> # ADM <dbl>, AGER <dbl>, AGR2 <dbl>, AGR3 <dbl>, AGRN <dbl>, AGRP <dbl>,
#> # AGXT <dbl>, AHCY <dbl>, AHSP <dbl>, AIF1 <dbl>, AIFM1 <dbl>, AK1 <dbl>, …
#>
#> $test_data
#> # A tibble: 118 × 102
#> DAid Age AARSD1 ABL1 ACAA1 ACAN ACE2 ACOX1 ACP5 ACP6 ACTA2
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 DA004… 81 2.47 0.793 -0.996 0.790 0.0125 -0.946 0.122 2.25 3.52
#> 2 DA005… 86 NA NA 0.441 0.185 -0.234 -1.26 1.30 0.792 0.491
#> 3 DA004… 51 3.63 2.89 3.73 0.481 0.319 0.712 1.48 1.90 3.60
#> 4 DA001… 47 3.14 1.77 NA -0.489 -0.780 1.41 1.40 1.79 -0.298
#> 5 DA001… 60 6.75 7.43 1.63 0.478 0.305 0.274 1.18 3.01 1.92
#> 6 DA000… 86 6.83 1.18 -1.74 -0.156 1.53 -0.721 0.620 0.527 0.772
#> 7 DA002… 71 2.97 2.73 2.66 1.03 0.471 0.636 -0.600 1.88 1.45
#> 8 DA003… 88 2.85 0.719 0.918 2.64 3.18 0.342 1.72 -2.39 0.614
#> 9 DA001… 86 3.13 1.91 -0.296 -0.0146 0.0954 -1.02 0.378 0.735 1.23
#> 10 DA005… 81 4.32 3.08 2.44 0.617 1.12 1.29 1.31 1.72 2.20
#> # ℹ 108 more rows
#> # ℹ 91 more variables: ACTN4 <dbl>, ACY1 <dbl>, ADA <dbl>, ADA2 <dbl>,
#> # ADAM15 <dbl>, ADAM23 <dbl>, ADAM8 <dbl>, ADAMTS13 <dbl>, ADAMTS15 <dbl>,
#> # ADAMTS16 <dbl>, ADAMTS8 <dbl>, ADCYAP1R1 <dbl>, ADGRE2 <dbl>, ADGRE5 <dbl>,
#> # ADGRG1 <dbl>, ADGRG2 <dbl>, ADH4 <dbl>, ADM <dbl>, AGER <dbl>, AGR2 <dbl>,
#> # AGR3 <dbl>, AGRN <dbl>, AGRP <dbl>, AGXT <dbl>, AHCY <dbl>, AHSP <dbl>,
#> # AIF1 <dbl>, AIFM1 <dbl>, AK1 <dbl>, AKR1B1 <dbl>, AKR1C4 <dbl>, …
#>
#> $model_type
#> [1] "regression"
#>
#> $final_workflow
#> ══ Workflow ════════════════════════════════════════════════════════════════════
#> Preprocessor: Recipe
#> Model: linear_reg()
#>
#> ── Preprocessor ────────────────────────────────────────────────────────────────
#> 5 Recipe Steps
#>
#> • step_dummy()
#> • step_nzv()
#> • step_normalize()
#> • step_corr()
#> • step_impute_knn()
#>
#> ── Model ───────────────────────────────────────────────────────────────────────
#> Linear Regression Model Specification (regression)
#>
#> Main Arguments:
#> penalty = 2.60336915648323e-09
#> mixture = 0.249163427995518
#>
#> Computational engine: glmnet
#>
#>
#> $metrics
#> $metrics$rmse
#> [1] 18.17391
#>
#> $metrics$rsq
#> [1] 0.02013586
#>
#>
#> $comparison_plot
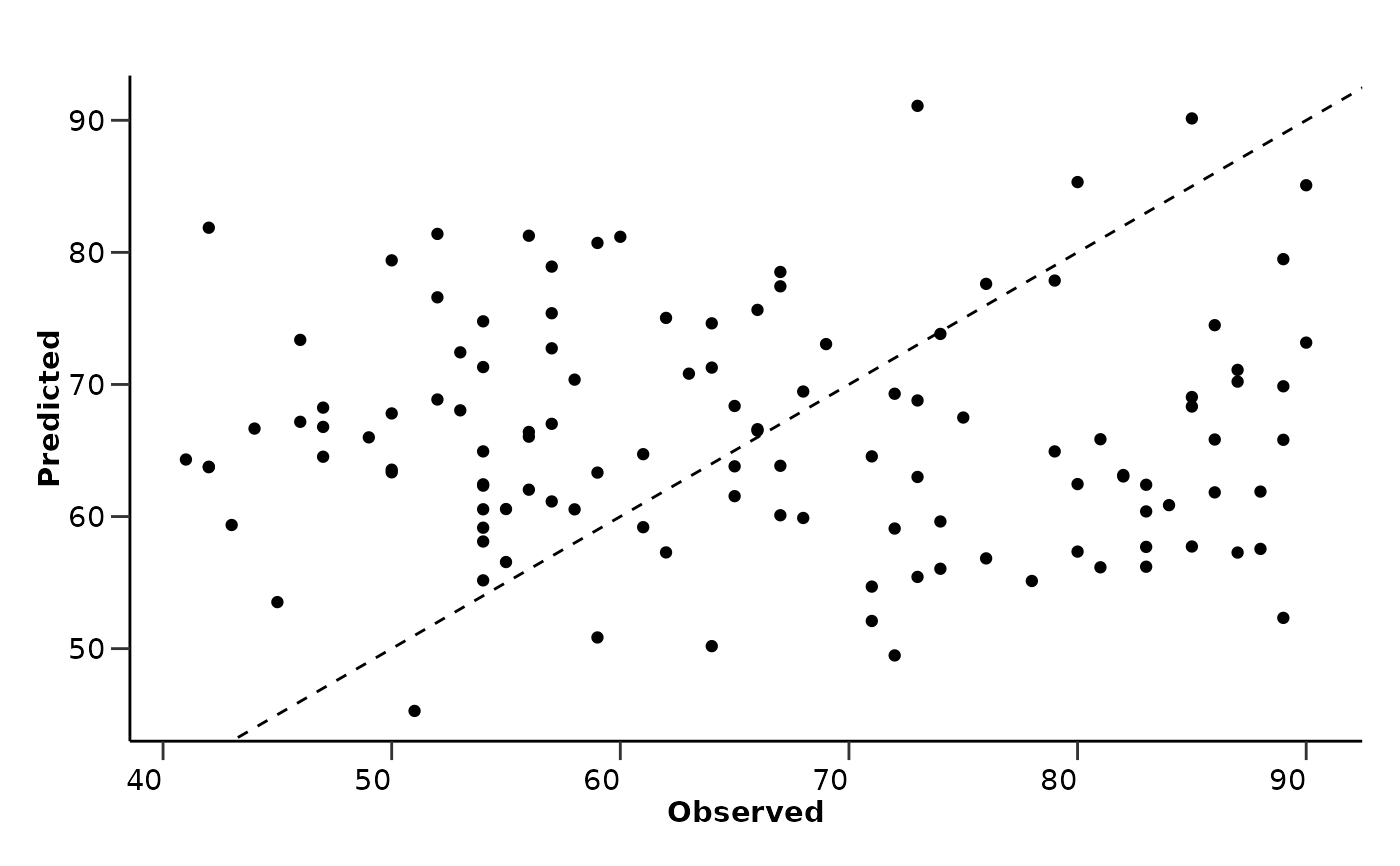 #>
#> $mixture
#> [1] 0.2491634
#>
#> $features
#> # A tibble: 100 × 4
#> Feature Importance Sign Scaled_Importance
#> <fct> <dbl> <chr> <dbl>
#> 1 ARID4B 2.87 POS 1
#> 2 ANGPT2 2.79 NEG 0.975
#> 3 ADAM15 2.57 POS 0.896
#> 4 AREG 2.54 POS 0.888
#> 5 ATP5IF1 2.52 NEG 0.880
#> 6 AHCY 2.36 POS 0.823
#> 7 B4GALT1 2.31 NEG 0.806
#> 8 ALDH1A1 2.31 POS 0.806
#> 9 AIFM1 1.94 POS 0.676
#> 10 ANXA11 1.86 POS 0.648
#> # ℹ 90 more rows
#>
#> $feat_imp_plot
#>
#> $mixture
#> [1] 0.2491634
#>
#> $features
#> # A tibble: 100 × 4
#> Feature Importance Sign Scaled_Importance
#> <fct> <dbl> <chr> <dbl>
#> 1 ARID4B 2.87 POS 1
#> 2 ANGPT2 2.79 NEG 0.975
#> 3 ADAM15 2.57 POS 0.896
#> 4 AREG 2.54 POS 0.888
#> 5 ATP5IF1 2.52 NEG 0.880
#> 6 AHCY 2.36 POS 0.823
#> 7 B4GALT1 2.31 NEG 0.806
#> 8 ALDH1A1 2.31 POS 0.806
#> 9 AIFM1 1.94 POS 0.676
#> 10 ANXA11 1.86 POS 0.648
#> # ℹ 90 more rows
#>
#> $feat_imp_plot
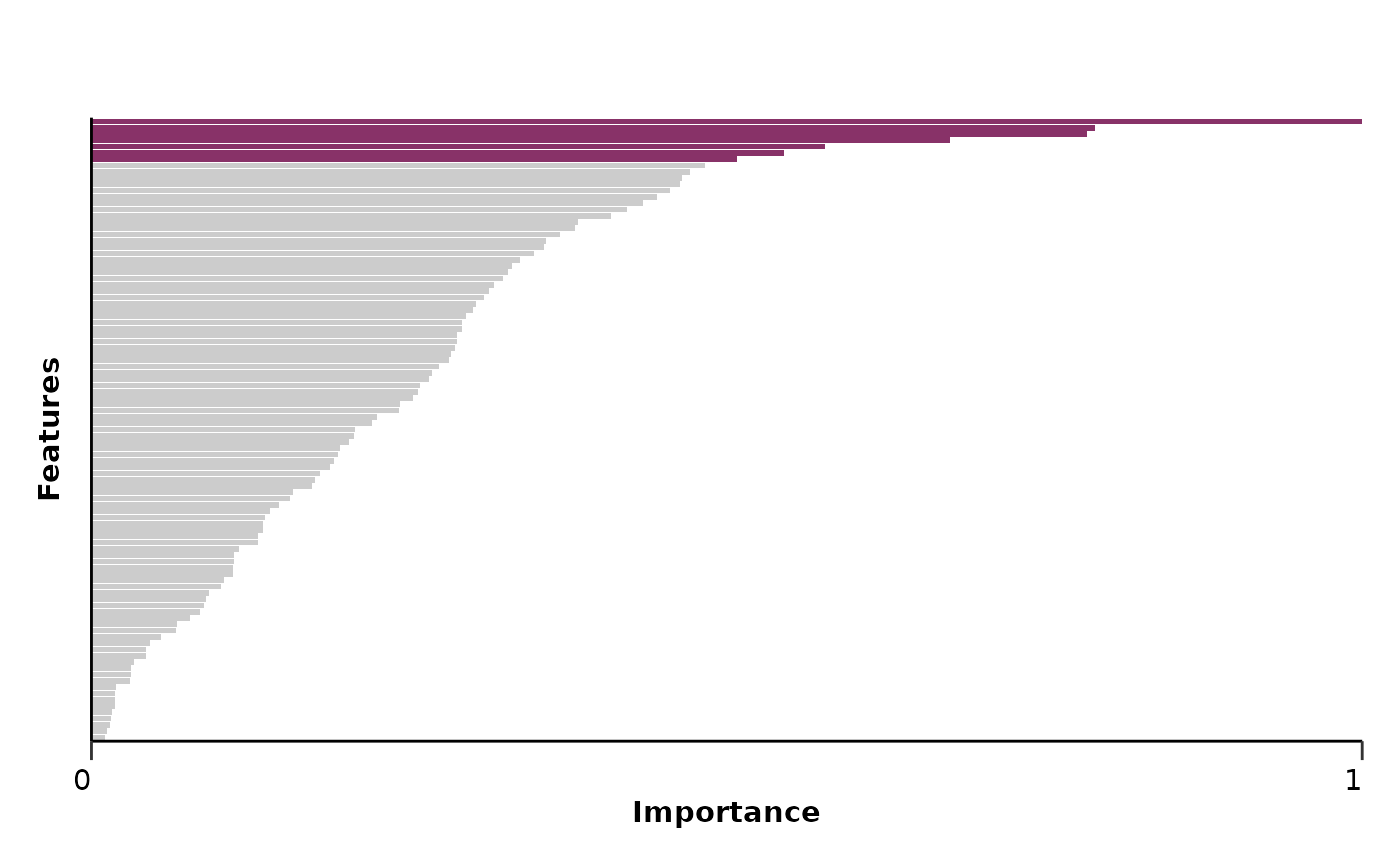 #>
#> $validation_data
#> # A tibble: 118 × 102
#> DAid Age AARSD1 ABL1 ACAA1 ACAN ACE2 ACOX1 ACP5 ACP6
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 DA00003 61 NA NA NA 0.989 NA 0.330 1.37 NA
#> 2 DA00007 85 NA NA 3.96 0.682 3.14 2.62 1.47 2.25
#> 3 DA00012 78 4.31 0.710 -1.44 -0.218 -0.469 -0.361 -0.0714 -1.30
#> 4 DA00014 68 6.34 7.25 5.12 0.0193 1.29 0.370 -0.382 0.830
#> 5 DA00016 78 1.79 1.36 0.106 -0.372 3.40 -1.19 1.77 1.07
#> 6 DA00030 67 3.31 5.38 4.82 0.266 0.606 3.12 1.22 2.13
#> 7 DA00032 62 3.62 3.06 -1.34 0.965 1.05 1.53 0.152 -0.124
#> 8 DA00038 69 2.23 1.42 0.484 1.72 1.46 0.0747 1.82 0.109
#> 9 DA00043 78 2.48 1.49 0.605 0.339 0.436 0.690 1.11 0.0158
#> 10 DA00051 82 2.53 3.00 0.166 0.707 -0.00699 1.05 0.898 1.53
#> # ℹ 108 more rows
#> # ℹ 92 more variables: ACTA2 <dbl>, ACTN4 <dbl>, ACY1 <dbl>, ADA <dbl>,
#> # ADA2 <dbl>, ADAM15 <dbl>, ADAM23 <dbl>, ADAM8 <dbl>, ADAMTS13 <dbl>,
#> # ADAMTS15 <dbl>, ADAMTS16 <dbl>, ADAMTS8 <dbl>, ADCYAP1R1 <dbl>,
#> # ADGRE2 <dbl>, ADGRE5 <dbl>, ADGRG1 <dbl>, ADGRG2 <dbl>, ADH4 <dbl>,
#> # ADM <dbl>, AGER <dbl>, AGR2 <dbl>, AGR3 <dbl>, AGRN <dbl>, AGRP <dbl>,
#> # AGXT <dbl>, AHCY <dbl>, AHSP <dbl>, AIF1 <dbl>, AIFM1 <dbl>, AK1 <dbl>, …
#>
#> $test_metrics
#> $test_metrics$rmse
#> [1] 16.58855
#>
#> $test_metrics$rsq
#> [1] 0.001185439
#>
#>
#> $test_comparison_plot
#>
#> $validation_data
#> # A tibble: 118 × 102
#> DAid Age AARSD1 ABL1 ACAA1 ACAN ACE2 ACOX1 ACP5 ACP6
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 DA00003 61 NA NA NA 0.989 NA 0.330 1.37 NA
#> 2 DA00007 85 NA NA 3.96 0.682 3.14 2.62 1.47 2.25
#> 3 DA00012 78 4.31 0.710 -1.44 -0.218 -0.469 -0.361 -0.0714 -1.30
#> 4 DA00014 68 6.34 7.25 5.12 0.0193 1.29 0.370 -0.382 0.830
#> 5 DA00016 78 1.79 1.36 0.106 -0.372 3.40 -1.19 1.77 1.07
#> 6 DA00030 67 3.31 5.38 4.82 0.266 0.606 3.12 1.22 2.13
#> 7 DA00032 62 3.62 3.06 -1.34 0.965 1.05 1.53 0.152 -0.124
#> 8 DA00038 69 2.23 1.42 0.484 1.72 1.46 0.0747 1.82 0.109
#> 9 DA00043 78 2.48 1.49 0.605 0.339 0.436 0.690 1.11 0.0158
#> 10 DA00051 82 2.53 3.00 0.166 0.707 -0.00699 1.05 0.898 1.53
#> # ℹ 108 more rows
#> # ℹ 92 more variables: ACTA2 <dbl>, ACTN4 <dbl>, ACY1 <dbl>, ADA <dbl>,
#> # ADA2 <dbl>, ADAM15 <dbl>, ADAM23 <dbl>, ADAM8 <dbl>, ADAMTS13 <dbl>,
#> # ADAMTS15 <dbl>, ADAMTS16 <dbl>, ADAMTS8 <dbl>, ADCYAP1R1 <dbl>,
#> # ADGRE2 <dbl>, ADGRE5 <dbl>, ADGRG1 <dbl>, ADGRG2 <dbl>, ADH4 <dbl>,
#> # ADM <dbl>, AGER <dbl>, AGR2 <dbl>, AGR3 <dbl>, AGRN <dbl>, AGRP <dbl>,
#> # AGXT <dbl>, AHCY <dbl>, AHSP <dbl>, AIF1 <dbl>, AIFM1 <dbl>, AK1 <dbl>, …
#>
#> $test_metrics
#> $test_metrics$rmse
#> [1] 16.58855
#>
#> $test_metrics$rsq
#> [1] 0.001185439
#>
#>
#> $test_comparison_plot
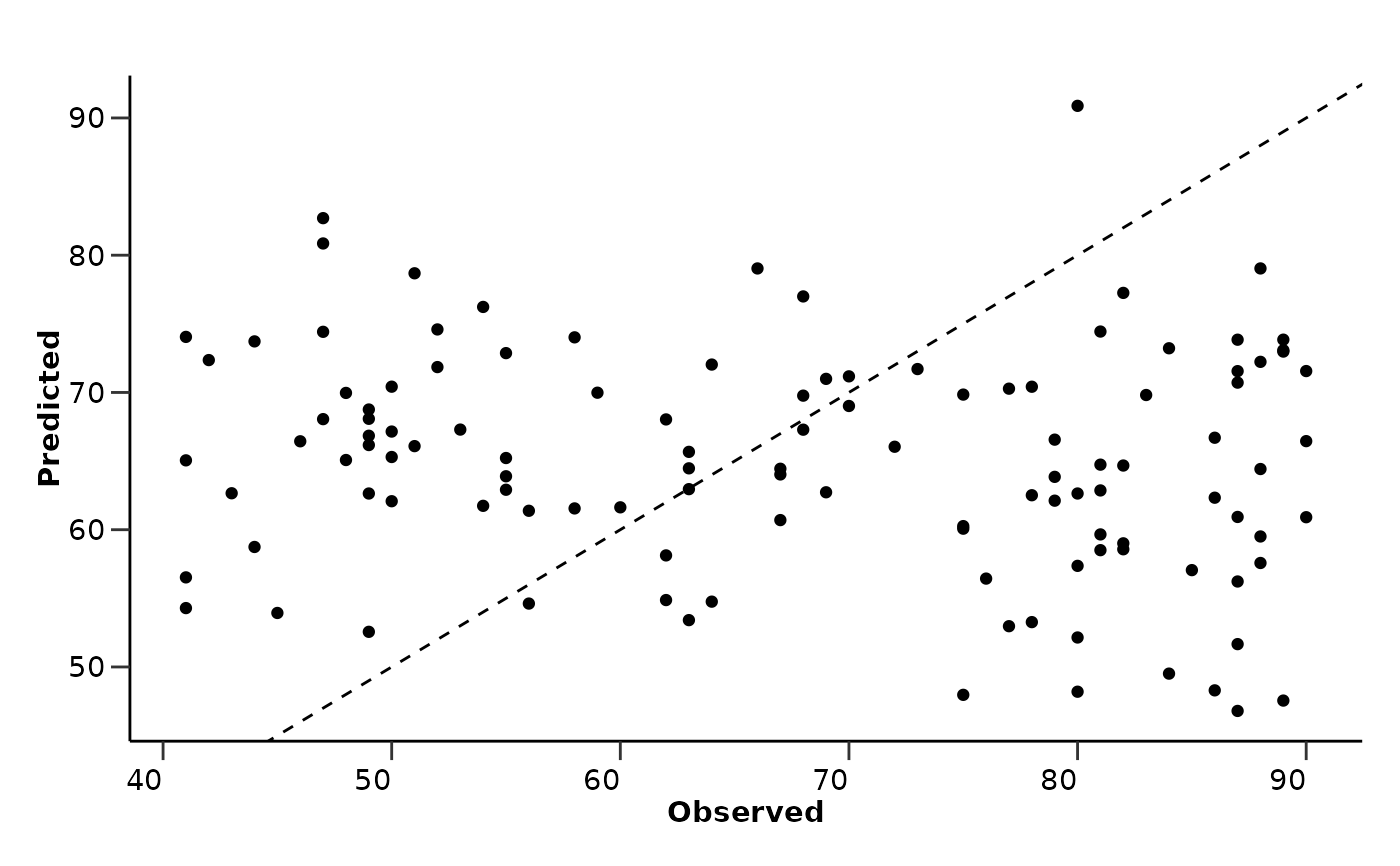 #>
#> attr(,"class")
#> [1] "hd_model"
#>
#> attr(,"class")
#> [1] "hd_model"
