na_search() provides a summary of missing values in a dataset. It allows the user to
specify the metadata columns to include in the summary and the color palette to use for
the heatmap annotations.
Usage
na_search(
olink_data,
metadata,
wide = TRUE,
metadata_cols = NULL,
palette = NULL,
x_labels = FALSE,
y_labels = FALSE,
show_heatmap = TRUE
)Arguments
- olink_data
The Olink dataset.
- metadata
The metadata dataset.
- wide
If TRUE, the data is in wide format.
- metadata_cols
The metadata columns to include in the summary.
- palette
The color palettes to use for the heatmap annotations (check examples bellow).
- x_labels
If TRUE, show x-axis labels.
- y_labels
If TRUE, show y-axis labels.
- show_heatmap
If TRUE, show the heatmap.
Details
When using continuous metadata variables, consider converted them to categorical by binning them into categories before passing them to the function. This will make the heatmap more informative and easier to interpret. Also when coloring annotations, the user can use custom palettes or the Human Protein Atlas (HPA) palettes. It is not required to provide a palette for all annotations, but when a palette is provided, it must be in correct format (check examples bellow).
Examples
# Use custom palettes for coloring annotations
palette = list(Sex = c(M = "blue", F = "pink"))
na_res <- na_search(example_data,
example_metadata,
wide = FALSE,
metadata_cols = c("Age", "Sex"),
palette = palette,
show_heatmap = FALSE)
# Use HPA palettes for coloring annotations
palette = list(Disease = get_hpa_palettes()$cancers12, Sex = get_hpa_palettes()$sex_hpa)
na_res <- na_search(example_data,
example_metadata,
wide = FALSE,
metadata_cols = c("Disease", "Sex"),
palette = palette,
show_heatmap = FALSE)
# Pre-bin a continuous variable
metadata <- example_metadata
metadata$Age_bin <- cut(metadata$Age,
breaks = c(0, 20, 40, 60, 80, 120),
labels = c("0-20", "21-40", "41-60", "61-80", "81+"),
right = FALSE)
palette = list(Disease = get_hpa_palettes()$cancers12)
na_search(example_data,
metadata,
wide = FALSE,
metadata_cols = c("Age_bin", "Disease"),
palette = palette)
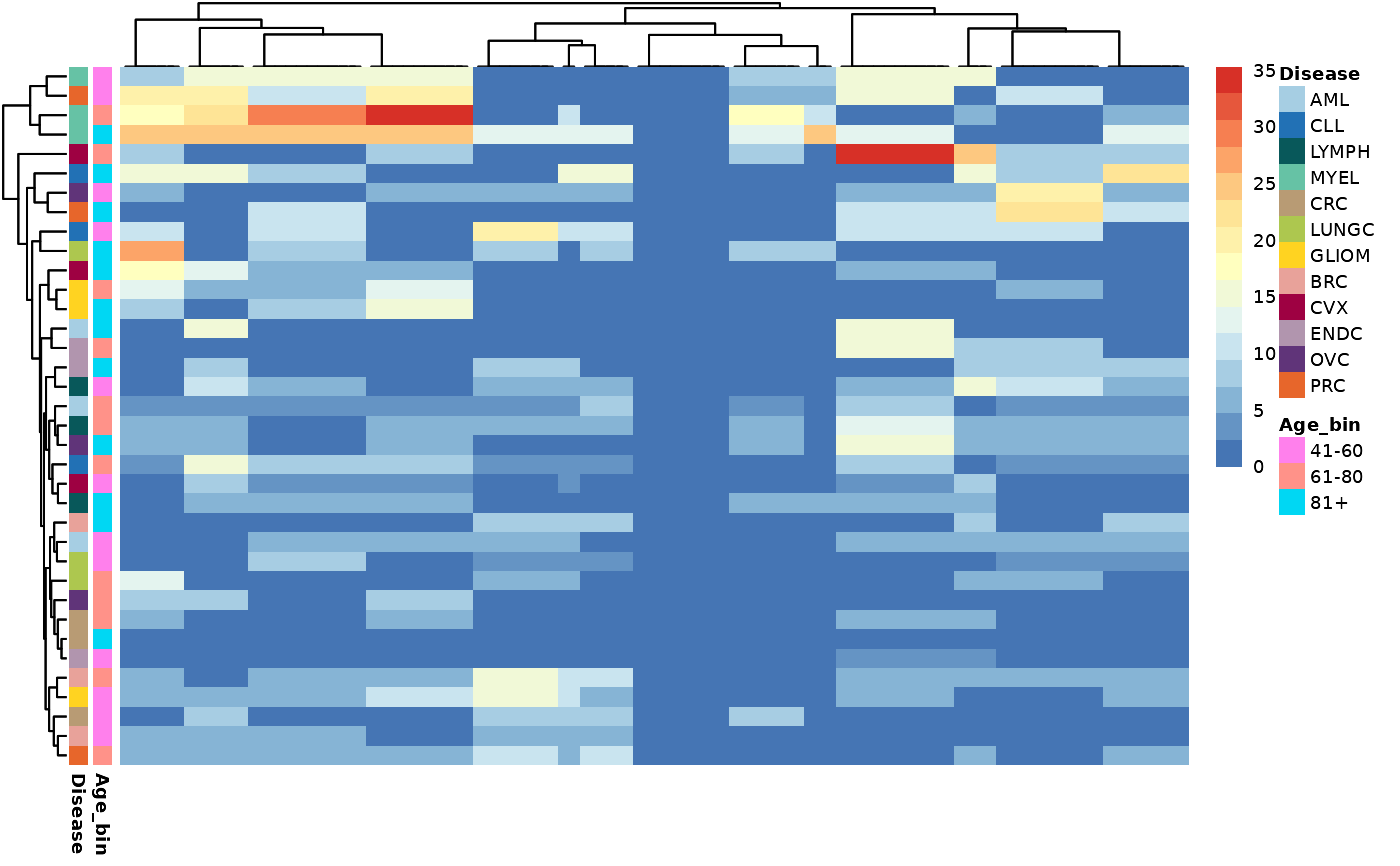 #> $na_data
#> # A tibble: 3,600 × 5
#> Age_bin Disease Categories Assay NA_percentage
#> <fct> <chr> <chr> <chr> <dbl>
#> 1 41-60 AML 41-60_AML AARSD1 5
#> 2 41-60 AML 41-60_AML ABL1 5
#> 3 41-60 AML 41-60_AML ACAA1 5
#> 4 41-60 AML 41-60_AML ACAN 5
#> 5 41-60 AML 41-60_AML ACE2 0
#> 6 41-60 AML 41-60_AML ACOX1 5
#> 7 41-60 AML 41-60_AML ACP5 0
#> 8 41-60 AML 41-60_AML ACP6 0
#> 9 41-60 AML 41-60_AML ACTA2 0
#> 10 41-60 AML 41-60_AML ACTN4 0
#> # ℹ 3,590 more rows
#>
#> $na_heatmap
#>
#> $na_data
#> # A tibble: 3,600 × 5
#> Age_bin Disease Categories Assay NA_percentage
#> <fct> <chr> <chr> <chr> <dbl>
#> 1 41-60 AML 41-60_AML AARSD1 5
#> 2 41-60 AML 41-60_AML ABL1 5
#> 3 41-60 AML 41-60_AML ACAA1 5
#> 4 41-60 AML 41-60_AML ACAN 5
#> 5 41-60 AML 41-60_AML ACE2 0
#> 6 41-60 AML 41-60_AML ACOX1 5
#> 7 41-60 AML 41-60_AML ACP5 0
#> 8 41-60 AML 41-60_AML ACP6 0
#> 9 41-60 AML 41-60_AML ACTA2 0
#> 10 41-60 AML 41-60_AML ACTN4 0
#> # ℹ 3,590 more rows
#>
#> $na_heatmap
#>
