do_xgboost_multi() runs the XGBoost multiclassification model pipeline. It splits the
data into training and test sets, creates class-balanced case-control groups,
and fits the model. It performs hyperparameter optimization and fits the best
model. It also plots the ROC curve and the AUC barplot for each class.
Usage
do_xgboost_multi(
olink_data,
metadata,
variable = "Disease",
wide = TRUE,
strata = TRUE,
exclude_cols = "Sex",
ratio = 0.75,
cor_threshold = 0.9,
normalize = TRUE,
cv_sets = 5,
grid_size = 50,
ncores = 4,
hypopt_vis = TRUE,
palette = NULL,
vline = TRUE,
varimp_yaxis_names = FALSE,
seed = 123
)Arguments
- olink_data
Olink data.
- metadata
Metadata.
- variable
The variable to predict. Default is "Disease".
- wide
Whether the data is wide format. Default is TRUE.
- strata
Whether to stratify the data. Default is TRUE.
- exclude_cols
Columns to exclude from the data before the model is tuned.
- ratio
Ratio of training data to test data. Default is 0.75.
- cor_threshold
Threshold of absolute correlation values. This will be used to remove the minimum number of features so that all their resulting absolute correlations are less than this value.
- normalize
Whether to normalize numeric data to have a standard deviation of one and a mean of zero. Default is TRUE.
- cv_sets
Number of cross-validation sets. Default is 5.
- grid_size
Size of the hyperparameter optimization grid. Default is 50.
- ncores
Number of cores to use for parallel processing. Default is 4.
- hypopt_vis
Whether to visualize hyperparameter optimization results. Default is TRUE.
- palette
The color palette for the plot. If it is a character, it should be one of the palettes from
get_hpa_palettes(). Default is NULL.- vline
Whether to add a vertical line at 50% importance. Default is TRUE.
- varimp_yaxis_names
Whether to add y-axis names to the variable importance plot. Default is FALSE.
- seed
Seed for reproducibility. Default is 123.
Value
A list with the following elements:
hypopt_res: Hyperparameter optimization results.
finalfit_res: Final model fitting results.
roc_curve: ROC curve plot.
auc: AUC values for each class.
auc_barplot: AUC barplot.
var_imp_res: Variable importance results.
Details
If the data contain missing values, KNN imputation will be applied.
If no check for feature correlation is preferred, set cor_threshold to 1.
It will filter out rows that contain NAs in Disease.
Examples
do_xgboost_multi(example_data,
example_metadata,
wide = FALSE,
palette = "cancers12",
cv_sets = 5,
grid_size = 5,
ncores = 1)
#> Joining with `by = join_by(DAid)`
#> Warning: Too little data to stratify.
#> • Resampling will be unstratified.
#> Sets are ready. Multiclassification model fitting is starting...
#> Warning: Too little data to stratify.
#> • Resampling will be unstratified.
#> Warning: Due to the small size of the grid, a Latin hypercube design will be used.
#> Warning: No event observations were detected in `truth` with event level 'BRC'.
#> Warning: No event observations were detected in `truth` with event level 'CLL'.
#> Warning: No event observations were detected in `truth` with event level 'CVX'.
#> Warning: No event observations were detected in `truth` with event level 'ENDC'.
#> Warning: No event observations were detected in `truth` with event level 'GLIOM'.
#> Warning: No event observations were detected in `truth` with event level 'LUNGC'.
#> Warning: No event observations were detected in `truth` with event level 'MYEL'.
#> Warning: No event observations were detected in `truth` with event level 'OVC'.
#> Warning: No event observations were detected in `truth` with event level 'AML'.
#> Warning: No event observations were detected in `truth` with event level 'CLL'.
#> Warning: No event observations were detected in `truth` with event level 'ENDC'.
#> Warning: No event observations were detected in `truth` with event level 'GLIOM'.
#> Warning: No event observations were detected in `truth` with event level 'LYMPH'.
#> Warning: No event observations were detected in `truth` with event level 'OVC'.
#> Warning: No event observations were detected in `truth` with event level 'CRC'.
#> Warning: No event observations were detected in `truth` with event level 'CVX'.
#> Warning: No event observations were detected in `truth` with event level 'ENDC'.
#> Warning: No event observations were detected in `truth` with event level 'GLIOM'.
#> Warning: No event observations were detected in `truth` with event level 'LUNGC'.
#> Warning: No event observations were detected in `truth` with event level 'LYMPH'.
#> Warning: No event observations were detected in `truth` with event level 'OVC'.
#> Warning: No event observations were detected in `truth` with event level 'AML'.
#> Warning: No event observations were detected in `truth` with event level 'CLL'.
#> Warning: No event observations were detected in `truth` with event level 'ENDC'.
#> Warning: No event observations were detected in `truth` with event level 'LYMPH'.
#> Warning: No event observations were detected in `truth` with event level 'MYEL'.
#> Warning: No event observations were detected in `truth` with event level 'AML'.
#> Warning: No event observations were detected in `truth` with event level 'CLL'.
#> Warning: No event observations were detected in `truth` with event level 'CRC'.
#> Warning: No event observations were detected in `truth` with event level 'GLIOM'.
#> Warning: No event observations were detected in `truth` with event level 'LUNGC'.
#> Warning: No event observations were detected in `truth` with event level 'MYEL'.
#> Warning: No event observations were detected in `truth` with event level 'AML'.
#> Warning: No event observations were detected in `truth` with event level 'CLL'.
#> Warning: No event observations were detected in `truth` with event level 'LUNGC'.
#> Warning: No event observations were detected in `truth` with event level 'LYMPH'.
#> Warning: No event observations were detected in `truth` with event level 'AML'.
#> Warning: No event observations were detected in `truth` with event level 'BRC'.
#> Warning: No event observations were detected in `truth` with event level 'CLL'.
#> Warning: No event observations were detected in `truth` with event level 'CRC'.
#> Warning: No event observations were detected in `truth` with event level 'CVX'.
#> Warning: No event observations were detected in `truth` with event level 'ENDC'.
#> Warning: No event observations were detected in `truth` with event level 'LUNGC'.
#> Warning: No event observations were detected in `truth` with event level 'MYEL'.
#> Warning: No event observations were detected in `truth` with event level 'PRC'.
#> Warning: No event observations were detected in `truth` with event level 'AML'.
#> Warning: No event observations were detected in `truth` with event level 'CLL'.
#> Warning: No event observations were detected in `truth` with event level 'CVX'.
#> Warning: No event observations were detected in `truth` with event level 'ENDC'.
#> Warning: No event observations were detected in `truth` with event level 'GLIOM'.
#> Warning: No event observations were detected in `truth` with event level 'MYEL'.
#> Warning: No event observations were detected in `truth` with event level 'PRC'.
#> Warning: No event observations were detected in `truth` with event level 'BRC'.
#> Warning: No event observations were detected in `truth` with event level 'CLL'.
#> Warning: No event observations were detected in `truth` with event level 'CRC'.
#> Warning: No event observations were detected in `truth` with event level 'GLIOM'.
#> Warning: No event observations were detected in `truth` with event level 'LUNGC'.
#> Warning: No event observations were detected in `truth` with event level 'MYEL'.
#> Warning: No event observations were detected in `truth` with event level 'OVC'.
#> Warning: No event observations were detected in `truth` with event level 'PRC'.
#> Warning: No event observations were detected in `truth` with event level 'BRC'.
#> Warning: No event observations were detected in `truth` with event level 'CLL'.
#> Warning: No event observations were detected in `truth` with event level 'LYMPH'.
#> Warning: No event observations were detected in `truth` with event level 'OVC'.
#> Warning: No event observations were detected in `truth` with event level 'PRC'.
#> Warning: No event observations were detected in `truth` with event level 'AML'.
#> Warning: No event observations were detected in `truth` with event level 'BRC'.
#> Warning: No event observations were detected in `truth` with event level 'CLL'.
#> Warning: No event observations were detected in `truth` with event level 'LYMPH'.
#> Warning: No event observations were detected in `truth` with event level 'MYEL'.
#> Warning: No event observations were detected in `truth` with event level 'AML'.
#> Warning: No event observations were detected in `truth` with event level 'CRC'.
#> Warning: No event observations were detected in `truth` with event level 'GLIOM'.
#> Warning: No event observations were detected in `truth` with event level 'MYEL'.
#> Warning: No event observations were detected in `truth` with event level 'OVC'.
#> $hypopt_res
#> $hypopt_res$xgboost_tune
#> # Tuning results
#> # 5-fold cross-validation using stratification
#> # A tibble: 5 × 5
#> splits id .metrics .notes .predictions
#> <list> <chr> <list> <list> <list>
#> 1 <split [351/88]> Fold1 <tibble [5 × 10]> <tibble [0 × 3]> <tibble [440 × 21]>
#> 2 <split [351/88]> Fold2 <tibble [5 × 10]> <tibble [0 × 3]> <tibble [440 × 21]>
#> 3 <split [351/88]> Fold3 <tibble [5 × 10]> <tibble [0 × 3]> <tibble [440 × 21]>
#> 4 <split [351/88]> Fold4 <tibble [5 × 10]> <tibble [0 × 3]> <tibble [440 × 21]>
#> 5 <split [352/87]> Fold5 <tibble [5 × 10]> <tibble [0 × 3]> <tibble [435 × 21]>
#>
#> $hypopt_res$xgboost_wf
#> ══ Workflow ════════════════════════════════════════════════════════════════════
#> Preprocessor: Recipe
#> Model: boost_tree()
#>
#> ── Preprocessor ────────────────────────────────────────────────────────────────
#> 4 Recipe Steps
#>
#> • step_normalize()
#> • step_nzv()
#> • step_corr()
#> • step_impute_knn()
#>
#> ── Model ───────────────────────────────────────────────────────────────────────
#> Boosted Tree Model Specification (classification)
#>
#> Main Arguments:
#> mtry = tune::tune()
#> trees = 1000
#> min_n = tune::tune()
#> tree_depth = tune::tune()
#> learn_rate = tune::tune()
#> loss_reduction = tune::tune()
#> sample_size = tune::tune()
#>
#> Computational engine: xgboost
#>
#>
#> $hypopt_res$train_set
#> # A tibble: 439 × 102
#> DAid AARSD1 ABL1 ACAA1 ACAN ACE2 ACOX1 ACP5 ACP6 ACTA2
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 DA00003 NA NA NA 0.989 NA 0.330 1.37 NA NA
#> 2 DA00004 3.41 3.38 1.69 NA 1.52 NA 0.841 0.582 1.70
#> 3 DA00005 5.01 5.05 0.128 0.401 -0.933 -0.584 0.0265 1.16 2.73
#> 4 DA00006 6.83 1.18 -1.74 -0.156 1.53 -0.721 0.620 0.527 0.772
#> 5 DA00007 NA NA 3.96 0.682 3.14 2.62 1.47 2.25 2.01
#> 6 DA00008 2.78 0.812 -0.552 0.982 -0.101 -0.304 0.376 -0.826 1.52
#> 7 DA00010 1.83 1.21 -0.912 -1.04 -0.0918 -0.304 1.69 0.0920 2.04
#> 8 DA00011 3.48 4.96 3.50 -0.338 4.48 1.26 2.18 1.62 1.79
#> 9 DA00012 4.31 0.710 -1.44 -0.218 -0.469 -0.361 -0.0714 -1.30 2.86
#> 10 DA00013 1.31 2.52 1.11 0.997 4.56 -1.35 0.833 2.33 3.57
#> # ℹ 429 more rows
#> # ℹ 92 more variables: ACTN4 <dbl>, ACY1 <dbl>, ADA <dbl>, ADA2 <dbl>,
#> # ADAM15 <dbl>, ADAM23 <dbl>, ADAM8 <dbl>, ADAMTS13 <dbl>, ADAMTS15 <dbl>,
#> # ADAMTS16 <dbl>, ADAMTS8 <dbl>, ADCYAP1R1 <dbl>, ADGRE2 <dbl>, ADGRE5 <dbl>,
#> # ADGRG1 <dbl>, ADGRG2 <dbl>, ADH4 <dbl>, ADM <dbl>, AGER <dbl>, AGR2 <dbl>,
#> # AGR3 <dbl>, AGRN <dbl>, AGRP <dbl>, AGXT <dbl>, AHCY <dbl>, AHSP <dbl>,
#> # AIF1 <dbl>, AIFM1 <dbl>, AK1 <dbl>, AKR1B1 <dbl>, AKR1C4 <dbl>, …
#>
#> $hypopt_res$test_set
#> # A tibble: 147 × 102
#> DAid AARSD1 ABL1 ACAA1 ACAN ACE2 ACOX1 ACP5 ACP6 ACTA2 ACTN4
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 DA00… 3.39 2.76 1.71 0.0333 1.76 -0.919 1.54 2.15 2.81 0.742
#> 2 DA00… 1.42 1.25 -0.816 -0.459 0.826 -0.902 0.647 1.30 0.798 -0.0659
#> 3 DA00… 4.39 3.34 -0.452 -0.868 0.395 1.71 1.49 -0.0285 0.200 -0.532
#> 4 DA00… 3.31 1.90 NA -0.926 0.408 0.687 1.03 0.612 2.19 0.258
#> 5 DA00… 1.46 0.832 -2.73 -0.371 2.27 0.0234 0.144 0.826 1.98 -0.280
#> 6 DA00… 2.62 2.48 0.537 -0.215 1.82 0.290 1.27 1.11 0.206 1.23
#> 7 DA00… 2.47 2.16 -0.486 NA 0.386 NA 1.38 0.536 1.86 0.00982
#> 8 DA00… 3.62 3.06 -1.34 0.965 1.05 1.53 0.152 -0.124 2.81 0.285
#> 9 DA00… 4.39 3.31 0.454 0.290 2.68 0.116 -1.32 0.945 2.14 -0.00881
#> 10 DA00… 0.964 2.94 1.55 1.67 2.50 0.164 1.83 1.46 3.03 0.449
#> # ℹ 137 more rows
#> # ℹ 91 more variables: ACY1 <dbl>, ADA <dbl>, ADA2 <dbl>, ADAM15 <dbl>,
#> # ADAM23 <dbl>, ADAM8 <dbl>, ADAMTS13 <dbl>, ADAMTS15 <dbl>, ADAMTS16 <dbl>,
#> # ADAMTS8 <dbl>, ADCYAP1R1 <dbl>, ADGRE2 <dbl>, ADGRE5 <dbl>, ADGRG1 <dbl>,
#> # ADGRG2 <dbl>, ADH4 <dbl>, ADM <dbl>, AGER <dbl>, AGR2 <dbl>, AGR3 <dbl>,
#> # AGRN <dbl>, AGRP <dbl>, AGXT <dbl>, AHCY <dbl>, AHSP <dbl>, AIF1 <dbl>,
#> # AIFM1 <dbl>, AK1 <dbl>, AKR1B1 <dbl>, AKR1C4 <dbl>, AKT1S1 <dbl>, …
#>
#> $hypopt_res$hypopt_vis
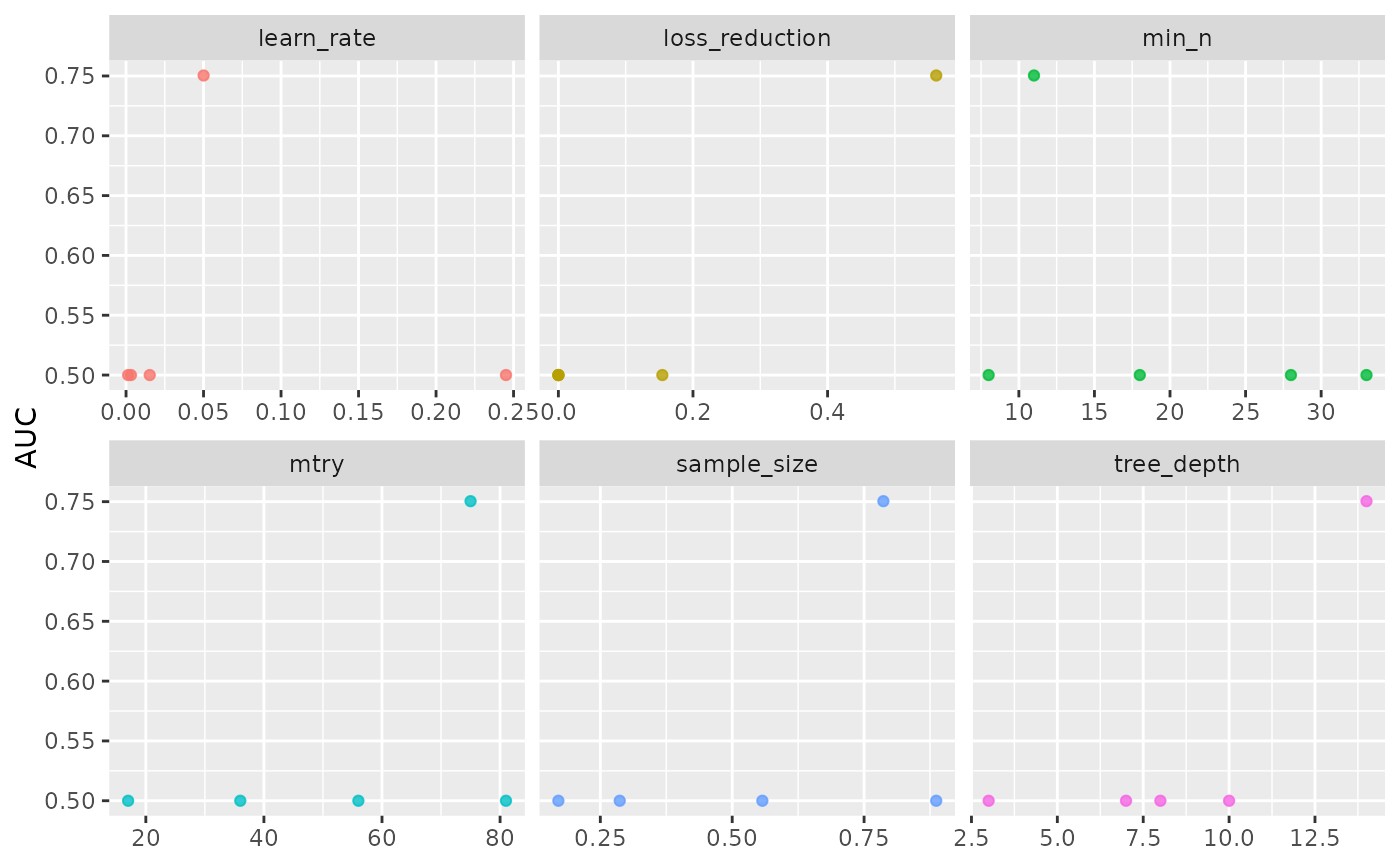 #>
#>
#> $finalfit_res
#> $finalfit_res$final
#> ══ Workflow [trained] ══════════════════════════════════════════════════════════
#> Preprocessor: Recipe
#> Model: boost_tree()
#>
#> ── Preprocessor ────────────────────────────────────────────────────────────────
#> 4 Recipe Steps
#>
#> • step_normalize()
#> • step_nzv()
#> • step_corr()
#> • step_impute_knn()
#>
#> ── Model ───────────────────────────────────────────────────────────────────────
#> ##### xgb.Booster
#> raw: 8.8 Mb
#> call:
#> xgboost::xgb.train(params = list(eta = 0.0500482848985379, max_depth = 14L,
#> gamma = 0.561465842160469, colsample_bytree = 1, colsample_bynode = 0.75,
#> min_child_weight = 11L, subsample = 0.786592039023526), data = x$data,
#> nrounds = 1000, watchlist = x$watchlist, verbose = 0, nthread = 1,
#> objective = "multi:softprob", num_class = 12L)
#> params (as set within xgb.train):
#> eta = "0.0500482848985379", max_depth = "14", gamma = "0.561465842160469", colsample_bytree = "1", colsample_bynode = "0.75", min_child_weight = "11", subsample = "0.786592039023526", nthread = "1", objective = "multi:softprob", num_class = "12", validate_parameters = "TRUE"
#> xgb.attributes:
#> niter
#> callbacks:
#> cb.evaluation.log()
#> # of features: 100
#> niter: 1000
#> nfeatures : 100
#> evaluation_log:
#> iter training_mlogloss
#> <num> <num>
#> 1 2.4575753
#> 2 2.4327717
#> --- ---
#> 999 0.3122689
#> 1000 0.3122298
#>
#> $finalfit_res$best
#> # A tibble: 1 × 6
#> mtry min_n tree_depth learn_rate loss_reduction sample_size
#> <int> <int> <int> <dbl> <dbl> <dbl>
#> 1 75 11 14 0.0500 0.561 0.787
#>
#> $finalfit_res$final_wf
#> ══ Workflow ════════════════════════════════════════════════════════════════════
#> Preprocessor: Recipe
#> Model: boost_tree()
#>
#> ── Preprocessor ────────────────────────────────────────────────────────────────
#> 4 Recipe Steps
#>
#> • step_normalize()
#> • step_nzv()
#> • step_corr()
#> • step_impute_knn()
#>
#> ── Model ───────────────────────────────────────────────────────────────────────
#> Boosted Tree Model Specification (classification)
#>
#> Main Arguments:
#> mtry = 75
#> trees = 1000
#> min_n = 11
#> tree_depth = 14
#> learn_rate = 0.0500482848985379
#> loss_reduction = 0.561465842160469
#> sample_size = 0.786592039023526
#>
#> Computational engine: xgboost
#>
#>
#>
#> $roc_curve
#>
#>
#> $finalfit_res
#> $finalfit_res$final
#> ══ Workflow [trained] ══════════════════════════════════════════════════════════
#> Preprocessor: Recipe
#> Model: boost_tree()
#>
#> ── Preprocessor ────────────────────────────────────────────────────────────────
#> 4 Recipe Steps
#>
#> • step_normalize()
#> • step_nzv()
#> • step_corr()
#> • step_impute_knn()
#>
#> ── Model ───────────────────────────────────────────────────────────────────────
#> ##### xgb.Booster
#> raw: 8.8 Mb
#> call:
#> xgboost::xgb.train(params = list(eta = 0.0500482848985379, max_depth = 14L,
#> gamma = 0.561465842160469, colsample_bytree = 1, colsample_bynode = 0.75,
#> min_child_weight = 11L, subsample = 0.786592039023526), data = x$data,
#> nrounds = 1000, watchlist = x$watchlist, verbose = 0, nthread = 1,
#> objective = "multi:softprob", num_class = 12L)
#> params (as set within xgb.train):
#> eta = "0.0500482848985379", max_depth = "14", gamma = "0.561465842160469", colsample_bytree = "1", colsample_bynode = "0.75", min_child_weight = "11", subsample = "0.786592039023526", nthread = "1", objective = "multi:softprob", num_class = "12", validate_parameters = "TRUE"
#> xgb.attributes:
#> niter
#> callbacks:
#> cb.evaluation.log()
#> # of features: 100
#> niter: 1000
#> nfeatures : 100
#> evaluation_log:
#> iter training_mlogloss
#> <num> <num>
#> 1 2.4575753
#> 2 2.4327717
#> --- ---
#> 999 0.3122689
#> 1000 0.3122298
#>
#> $finalfit_res$best
#> # A tibble: 1 × 6
#> mtry min_n tree_depth learn_rate loss_reduction sample_size
#> <int> <int> <int> <dbl> <dbl> <dbl>
#> 1 75 11 14 0.0500 0.561 0.787
#>
#> $finalfit_res$final_wf
#> ══ Workflow ════════════════════════════════════════════════════════════════════
#> Preprocessor: Recipe
#> Model: boost_tree()
#>
#> ── Preprocessor ────────────────────────────────────────────────────────────────
#> 4 Recipe Steps
#>
#> • step_normalize()
#> • step_nzv()
#> • step_corr()
#> • step_impute_knn()
#>
#> ── Model ───────────────────────────────────────────────────────────────────────
#> Boosted Tree Model Specification (classification)
#>
#> Main Arguments:
#> mtry = 75
#> trees = 1000
#> min_n = 11
#> tree_depth = 14
#> learn_rate = 0.0500482848985379
#> loss_reduction = 0.561465842160469
#> sample_size = 0.786592039023526
#>
#> Computational engine: xgboost
#>
#>
#>
#> $roc_curve
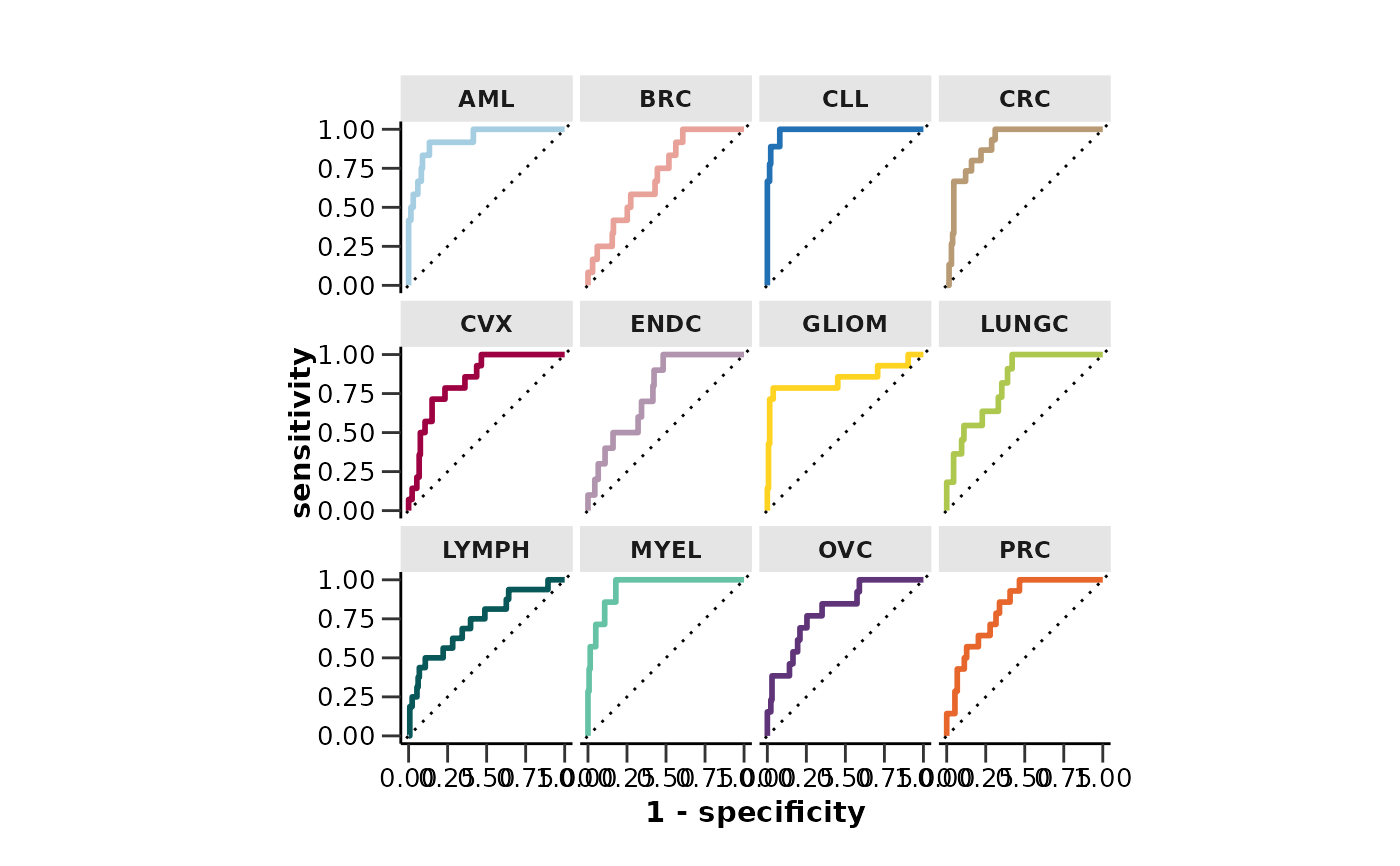 #>
#> $auc
#> # A tibble: 12 × 2
#> Disease AUC
#> <chr> <dbl>
#> 1 AML 0.841
#> 2 BRC 0.687
#> 3 CLL 0.881
#> 4 CRC 0.635
#> 5 CVX 0.697
#> 6 ENDC 0.715
#> 7 GLIOM 0.672
#> 8 LUNGC 0.622
#> 9 LYMPH 0.794
#> 10 MYEL 0.827
#> 11 OVC 0.740
#> 12 PRC 0.703
#>
#> $auc_barplot
#>
#> $auc
#> # A tibble: 12 × 2
#> Disease AUC
#> <chr> <dbl>
#> 1 AML 0.841
#> 2 BRC 0.687
#> 3 CLL 0.881
#> 4 CRC 0.635
#> 5 CVX 0.697
#> 6 ENDC 0.715
#> 7 GLIOM 0.672
#> 8 LUNGC 0.622
#> 9 LYMPH 0.794
#> 10 MYEL 0.827
#> 11 OVC 0.740
#> 12 PRC 0.703
#>
#> $auc_barplot
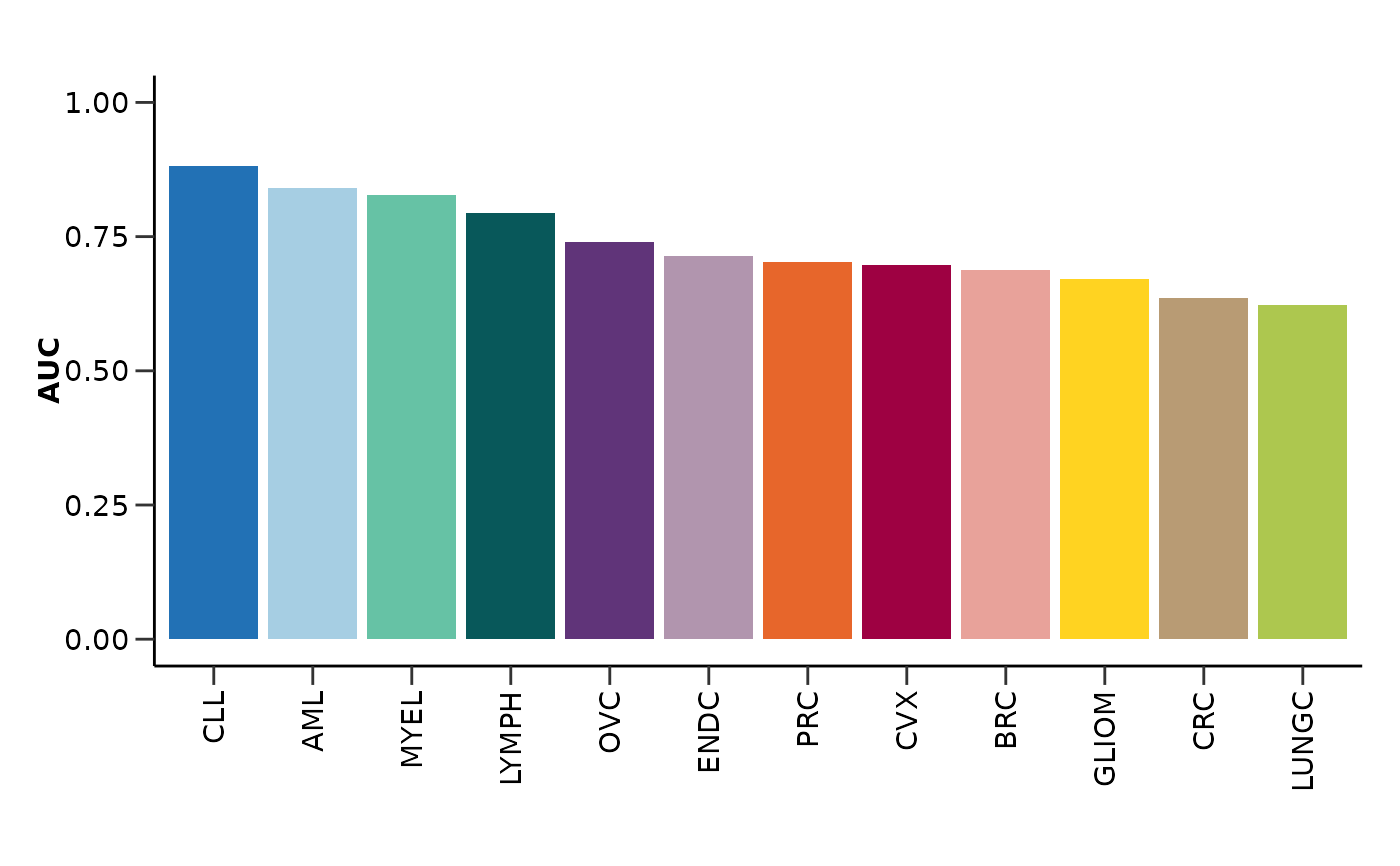 #>
#> $var_imp_res
#> $var_imp_res$features
#> # A tibble: 99 × 3
#> Variable Importance Scaled_Importance
#> <fct> <dbl> <dbl>
#> 1 APEX1 0.0305 100
#> 2 ALPP 0.0274 89.0
#> 3 ADAMTS15 0.0259 83.6
#> 4 ADA 0.0248 79.7
#> 5 ARID4B 0.0242 77.6
#> 6 AZU1 0.0241 77.3
#> 7 AHCY 0.0220 69.8
#> 8 ADAMTS16 0.0218 68.8
#> 9 ADGRG2 0.0209 65.6
#> 10 ACY1 0.0205 64.5
#> # ℹ 89 more rows
#>
#> $var_imp_res$var_imp_plot
#>
#> $var_imp_res
#> $var_imp_res$features
#> # A tibble: 99 × 3
#> Variable Importance Scaled_Importance
#> <fct> <dbl> <dbl>
#> 1 APEX1 0.0305 100
#> 2 ALPP 0.0274 89.0
#> 3 ADAMTS15 0.0259 83.6
#> 4 ADA 0.0248 79.7
#> 5 ARID4B 0.0242 77.6
#> 6 AZU1 0.0241 77.3
#> 7 AHCY 0.0220 69.8
#> 8 ADAMTS16 0.0218 68.8
#> 9 ADGRG2 0.0209 65.6
#> 10 ACY1 0.0205 64.5
#> # ℹ 89 more rows
#>
#> $var_imp_res$var_imp_plot
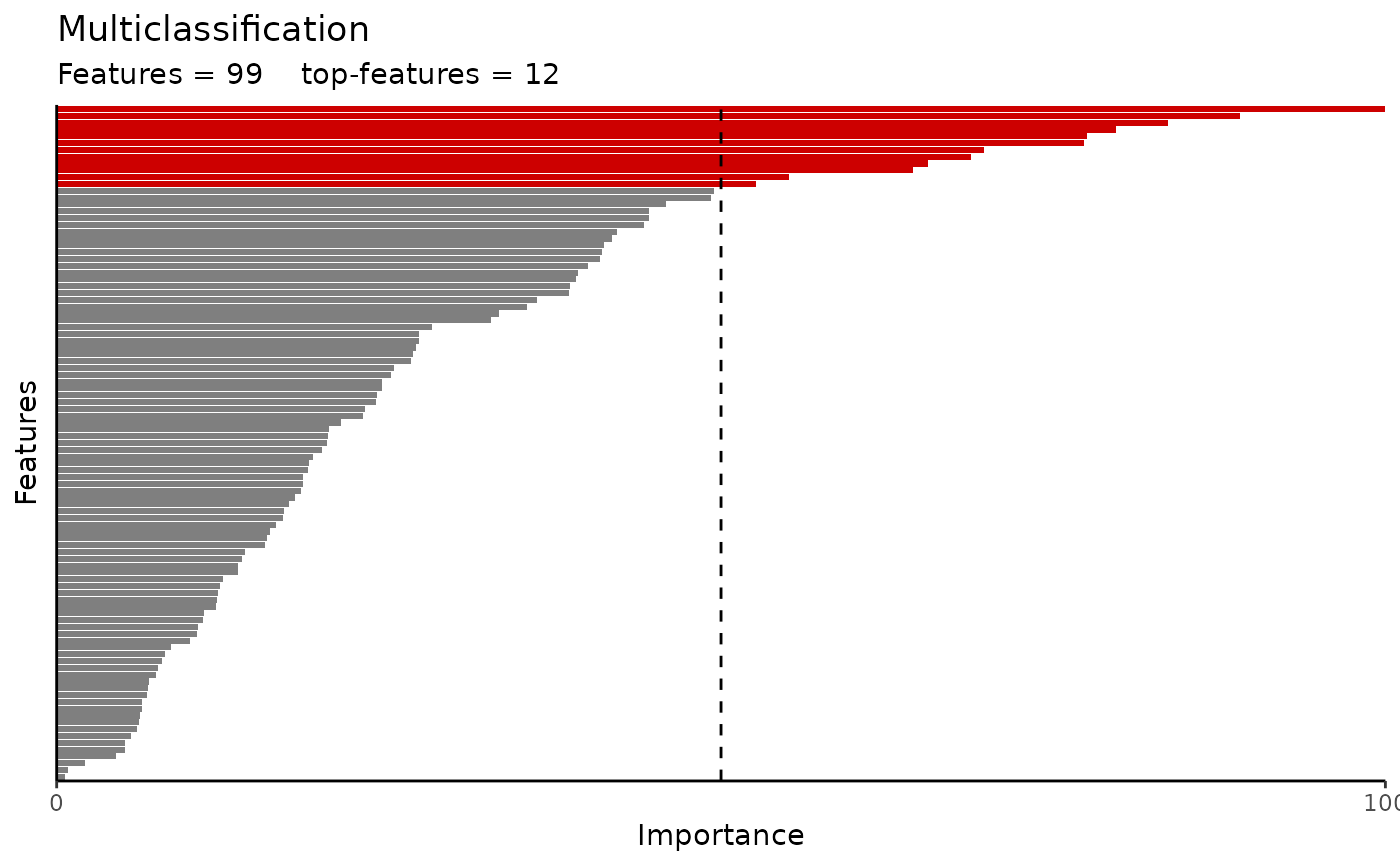 #>
#>
#>
#>
