
Run differential expression analysis with limma for continuous variable
Source:R/diff_expression.R
do_limma_continuous.RdThis function runs differential expression analysis using limma for a continuous variable. It can generate and save volcano plots.
Usage
do_limma_continuous(
olink_data,
metadata,
variable,
correct = c("Sex"),
correct_type = c("factor"),
wide = TRUE,
volcano = TRUE,
pval_lim = 0.05,
logfc_lim = 0,
top_up_prot = 40,
top_down_prot = 10,
palette = "diff_exp",
report_nproteins = TRUE,
user_defined_proteins = NULL,
subtitle = NULL,
save = FALSE
)Arguments
- olink_data
A tibble with the Olink data in wide format.
- metadata
A tibble with the metadata.
- variable
The variable of interest.
- correct
The variables to correct the results with. Default is c("Sex").
- correct_type
The type of the variables to correct the results with. Default is c("factor").
- wide
If the data is in wide format. Default is TRUE.
- volcano
Generate volcano plots. Default is TRUE.
- pval_lim
The p-value limit for significance. Default is 0.05.
- logfc_lim
The logFC limit for significance. Default is 0.
- top_up_prot
The number of top up regulated proteins to label on the plot. Default is 40.
- top_down_prot
The number of top down regulated proteins to label on the plot. Default is 10.
- palette
The color palette for the plot. If it is a character, it should be one of the palettes from
get_hpa_palettes(). Default is "diff_exp".- report_nproteins
If the number of significant proteins should be reported in the subtitle. Default is TRUE.
- user_defined_proteins
A list with the user defined proteins to label on the plot. Default is NULL.
- subtitle
The subtitle of the plot or NULL for no subtitle.
- save
Save the volcano plots. Default is FALSE.
Value
A list with the differential expression results and volcano plots.
de_results: A list with the differential expression results.
volcano_plot: A list with the volcano plots.
Details
It will filter out rows with NA values in any of the columns that are used for
correction, either the variable or in correct. The user_defined_proteins overrides
the top_up_prot and top_down_prot arguments.
Examples
do_limma_continuous(example_data, example_metadata, "Age", wide = FALSE)
#> $de_results
#> # A tibble: 100 × 9
#> Assay logFC as.factor.Sex.F as.factor.Sex.M AveExpr F P.Value
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 ADAMTS15 -0.000719 3.09 2.92 2.99 1874. 2.10e-291
#> 2 AARSD1 0.000327 2.96 3.25 3.13 1608. 1.29e-274
#> 3 AKT1S1 0.00154 3.28 3.46 3.47 1478. 3.54e-265
#> 4 ATG4A -0.00157 2.56 2.71 2.55 1138. 2.26e-238
#> 5 ATOX1 -0.00166 3.02 3.18 2.97 1061. 1.13e-232
#> 6 ADM 0.00536 1.53 1.47 1.87 954. 7.63e-224
#> 7 AK1 -0.00373 2.51 2.66 2.34 786. 3.94e-202
#> 8 AKR1B1 -0.000171 2.28 2.33 2.29 783. 2.64e-200
#> 9 ATP5IF1 -0.00321 3.66 4.02 3.60 740. 1.11e-196
#> 10 ARHGEF12 -0.00163 3.19 3.56 3.26 683. 2.06e-187
#> # ℹ 90 more rows
#> # ℹ 2 more variables: adj.P.Val <dbl>, sig <chr>
#>
#> $volcano_plot
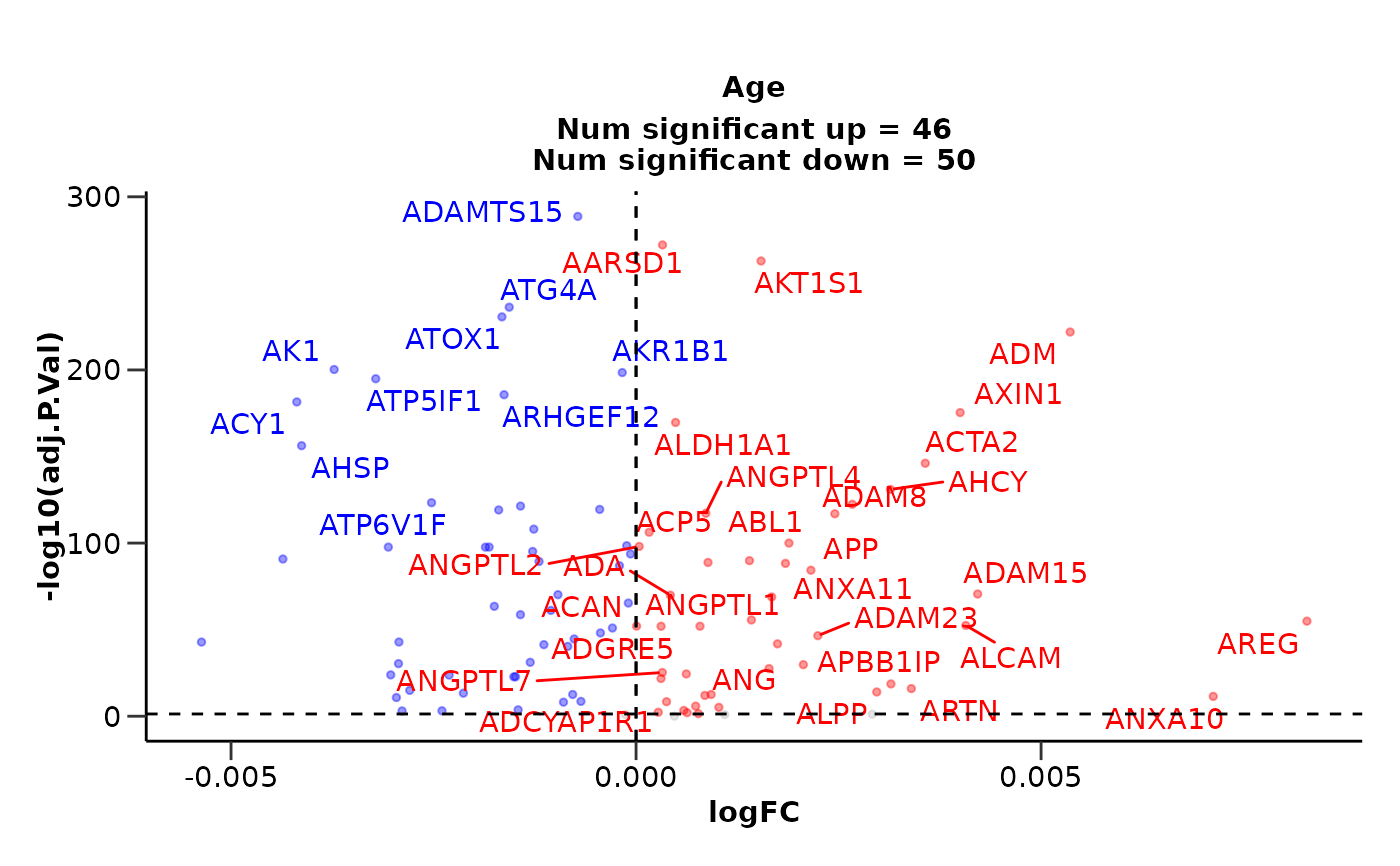 #>
#>