hd_wgcna() performs a weighted gene co-expression network analysis (WGCNA) on the provided data.
The user can specify the power parameter for the analysis or the function will select an optimal
power value based on the data. The function returns a list containing the WGCNA object, the power
and the power plots (in case optimization is performed). If the data contain missing values, the function
imputes them using the k-nearest neighbors algorithm (k = 5).
Details
If you want to learn more about WGCNA, please refer to the following tutorial:
https://edo98811.github.io/WGCNA_official_documentation/
Examples
# Initialize an HDAnalyzeR object
hd_object <- hd_initialize(example_data, example_metadata)
# Perform WGCNA analysis
wgcna_res <- hd_wgcna(hd_object)
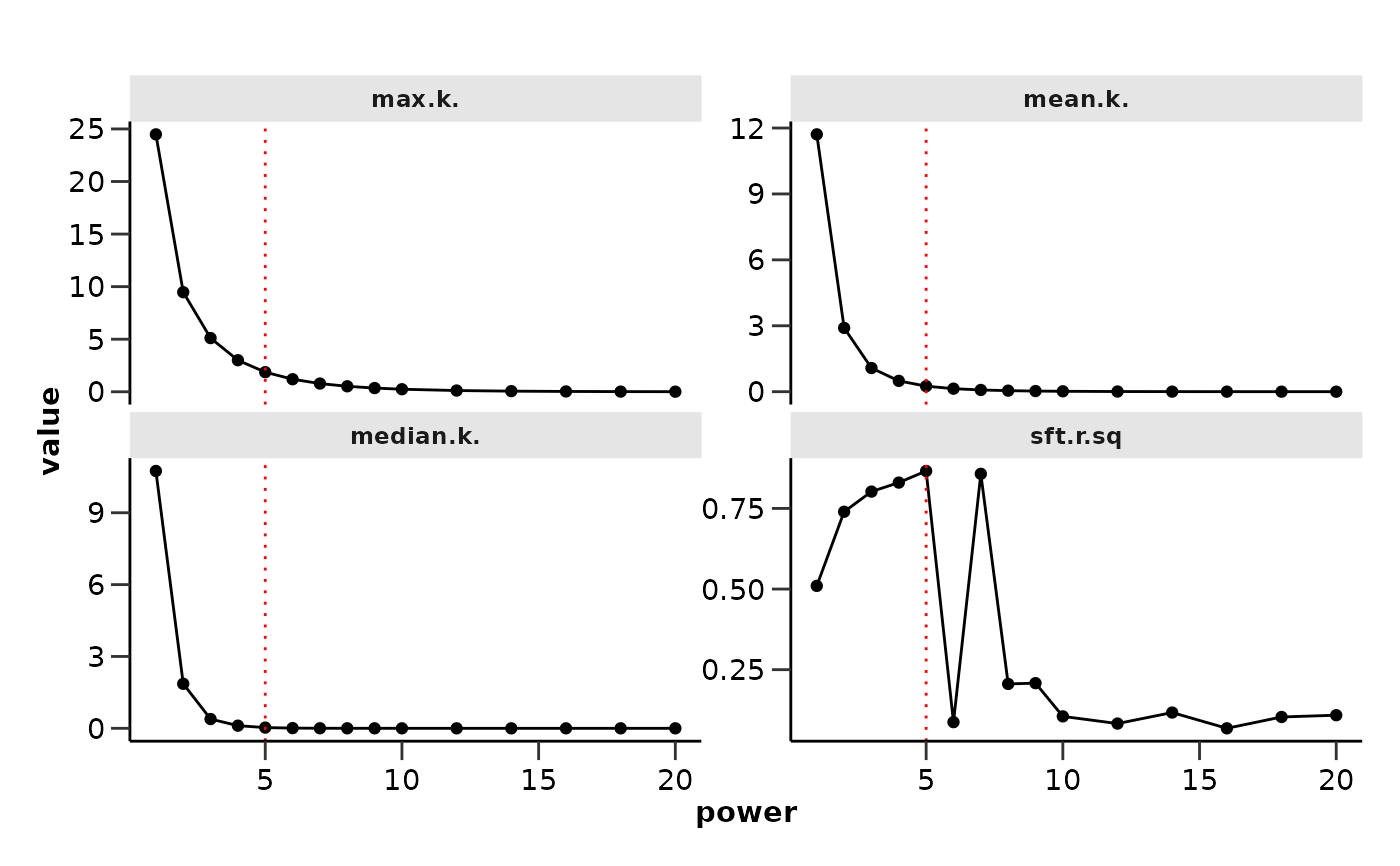
#> Power SFT.R.sq slope truncated.R.sq mean.k. median.k. max.k.
#> 1 1 0.5100 -0.742 0.7080 1.17e+01 1.07e+01 24.50000
#> 2 2 0.7400 -0.863 0.7370 2.90e+00 1.86e+00 9.47000
#> 3 3 0.8020 -0.876 0.8210 1.08e+00 3.88e-01 5.11000
#> 4 4 0.8300 -0.882 0.8000 4.92e-01 1.11e-01 3.00000
#> 5 5 0.8660 -0.947 0.8340 2.51e-01 3.05e-02 1.86000
#> 6 6 0.0873 -1.240 -0.1180 1.37e-01 9.13e-03 1.19000
#> 7 7 0.8570 -0.979 0.8460 7.87e-02 2.87e-03 0.78100
#> 8 8 0.2060 -1.870 0.0826 4.67e-02 9.18e-04 0.52100
#> 9 9 0.2080 -1.830 0.0607 2.85e-02 3.02e-04 0.35300
#> 10 10 0.1050 -1.250 -0.0824 1.78e-02 1.01e-04 0.24200
#> 11 12 0.0828 -1.550 -0.0219 7.34e-03 1.18e-05 0.11700
#> 12 14 0.1170 -1.750 -0.0305 3.23e-03 1.50e-06 0.05900
#> 13 16 0.0681 -1.520 0.0990 1.50e-03 1.98e-07 0.03040
#> 14 18 0.1030 -1.740 0.0846 7.24e-04 2.50e-08 0.01600
#> 15 20 0.1090 -1.720 0.0970 3.62e-04 3.22e-09 0.00853
#> mergeCloseModules: less than two proper modules.
#> ..color levels are grey, turquoise
#> ..there is nothing to merge.
# Access the WGCNA results
wgcna_res$wgcna$colors
#> AARSD1 ABL1 ACAA1 ACAN ACE2 ACOX1
#> "turquoise" "turquoise" "turquoise" "grey" "grey" "turquoise"
#> ACP5 ACP6 ACTA2 ACTN4 ACY1 ADA
#> "grey" "grey" "grey" "grey" "grey" "grey"
#> ADA2 ADAM15 ADAM23 ADAM8 ADAMTS13 ADAMTS15
#> "grey" "grey" "grey" "grey" "grey" "grey"
#> ADAMTS16 ADAMTS8 ADCYAP1R1 ADGRE2 ADGRE5 ADGRG1
#> "grey" "grey" "grey" "grey" "grey" "grey"
#> ADGRG2 ADH4 ADM AGER AGR2 AGR3
#> "grey" "grey" "grey" "grey" "grey" "grey"
#> AGRN AGRP AGXT AHCY AHSP AIF1
#> "grey" "grey" "grey" "turquoise" "grey" "turquoise"
#> AIFM1 AK1 AKR1B1 AKR1C4 AKT1S1 AKT3
#> "turquoise" "turquoise" "turquoise" "grey" "turquoise" "turquoise"
#> ALCAM ALDH1A1 ALDH3A1 ALPP AMBN AMBP
#> "grey" "grey" "grey" "grey" "grey" "grey"
#> AMFR AMIGO2 AMN AMY2A AMY2B ANG
#> "grey" "grey" "grey" "grey" "grey" "grey"
#> ANGPT1 ANGPT2 ANGPTL1 ANGPTL2 ANGPTL3 ANGPTL4
#> "turquoise" "grey" "grey" "grey" "grey" "grey"
#> ANGPTL7 ANKRD54 ANPEP ANXA10 ANXA11 ANXA3
#> "grey" "grey" "grey" "grey" "turquoise" "turquoise"
#> ANXA4 ANXA5 AOC1 AOC3 APBB1IP APEX1
#> "turquoise" "grey" "grey" "grey" "turquoise" "turquoise"
#> APLP1 APOH APOM APP AREG ARG1
#> "grey" "grey" "grey" "turquoise" "grey" "grey"
#> ARHGAP1 ARHGAP25 ARHGEF12 ARID4B ARNT ARSA
#> "turquoise" "turquoise" "turquoise" "grey" "grey" "grey"
#> ARSB ART3 ARTN ATF2 ATG4A ATOX1
#> "turquoise" "grey" "grey" "grey" "turquoise" "turquoise"
#> ATP5IF1 ATP5PO ATP6AP2 ATP6V1D ATP6V1F ATXN10
#> "turquoise" "grey" "grey" "grey" "turquoise" "turquoise"
#> AXIN1 AXL AZU1 B4GALT1
#> "turquoise" "grey" "grey" "grey"
head(wgcna_res$wgcna$MEs)
#> MEturquoise MEgrey
#> DA00001 0.01303185 0.079182698
#> DA00002 -0.06147570 0.001922882
#> DA00003 0.05582282 -0.040884118
#> DA00004 0.04934369 0.026582974
#> DA00005 0.03279477 0.065722451
#> DA00006 -0.00363437 -0.013683113
wgcna_res$power
#> [1] 5
wgcna_res$power_plots